1TG0
 
 | |
2O9V
 
 | |
1SSH
 
 | | Crystal structure of the SH3 domain from a S. cerevisiae hypothetical 40.4 kDa protein in complex with a peptide | | 分子名称: | 12-mer peptide from Cytoskeleton assembly control protein SLA1, Hypothetical 40.4 kDa protein in PES4-HIS2 intergenic region | | 著者 | Kursula, P, Kursula, I, Lehmann, F, Song, Y.-H, Wilmanns, M. | | 登録日 | 2004-03-24 | | 公開日 | 2005-04-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Yeast SH3 domain structural genomics
To be Published
|
|
2O31
 
 | |
2O9S
 
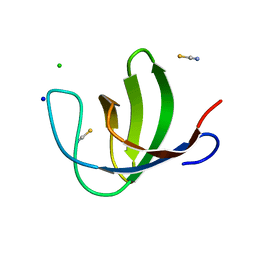 | |
5CGY
 
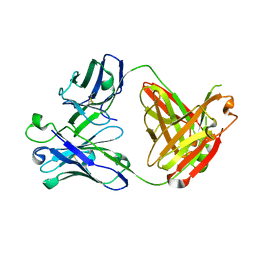 | |
8BEB
 
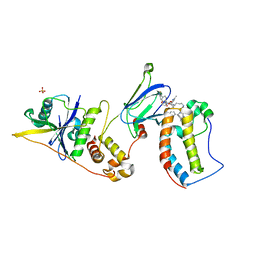 | | Ternary complex between VCB, BRD4-BD1 and PROTAC 49 | | 分子名称: | (2~{S},4~{R})-~{N}-[(1~{S})-3-[4-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]butylamino]-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-3-oxidanylidene-propyl]-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Elongin-B, ... | | 著者 | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | 登録日 | 2022-10-21 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
8BDX
 
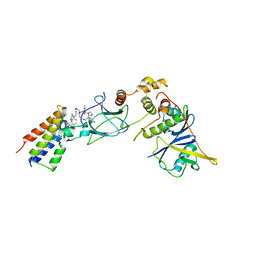 | | Ternary complex between VCB, BRD4-BD2 and PROTAC 48 | | 分子名称: | (2S,4R)-N-[(1S)-1-(4-chlorophenyl)-3-[2-[2-[2-[2-[2-[(9S)-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8$l^{5},11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethylamino]-3-oxidanylidene-propyl]-1-[(2R)-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Elongin-B, ... | | 著者 | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | 登録日 | 2022-10-20 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
5CHN
 
 | |
8BRF
 
 | |
8BS0
 
 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at 80 MPa helium gas pressure in a sapphire capillary | | 分子名称: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | 著者 | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | 登録日 | 2022-11-24 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BRZ
 
 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at 52 MPa helium gas pressure in a sapphire capillary | | 分子名称: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | 著者 | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | 登録日 | 2022-11-24 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BRY
 
 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at atmospheric pressure | | 分子名称: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | 著者 | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | 登録日 | 2022-11-24 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
4PF4
 
 | | 1.1A X-RAY STRUCTURE OF THE APO CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE 1, aa 1-277 | | 分子名称: | Death-associated protein kinase 1, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Temmerman, K, Simon, B, Wilmanns, M. | | 登録日 | 2014-04-28 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | 1.1A X-RAY STRUCTURE OF THE APO CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE 1, aa 1-277
To Be Published
|
|
4KAO
 
 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH 1-(5-tert-Butyl-2-p-tolyl-2H-pyrazol-3-yl)-3-(4-pyridin-3- yl-phenyl)-urea | | 分子名称: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[4-(pyridin-3-yl)phenyl]urea, Focal adhesion kinase 1, SULFATE ION | | 著者 | Musil, D, Graedler, U, Heinrich, T, Lehmann, M, Dresing, V. | | 登録日 | 2013-04-22 | | 公開日 | 2013-09-11 | | 最終更新日 | 2013-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Fragment-based discovery of focal adhesion kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8H2I
 
 | | Near-atomic structure of five-fold averaged PBCV-1 capsid | | 分子名称: | MCPv1, MCPv2, MCPv3, ... | | 著者 | Shao, Q, Agarkova, I.V, Noel, E.A, Dunigan, D.D, Liu, Y, Wang, A, Guo, M, Xie, L, Zhao, X, Rossmann, M.G, Van Etten, J.L, Klose, T, Fang, Q. | | 登録日 | 2022-10-06 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Near-atomic, non-icosahedrally averaged structure of giant virus Paramecium bursaria chlorella virus 1.
Nat Commun, 13, 2022
|
|
6QMO
 
 | |
1H1C
 
 | |
5CW2
 
 | |
6HXG
 
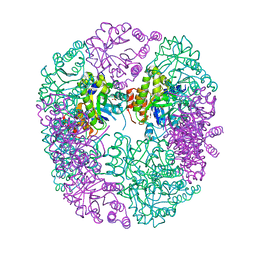 | | PDX1.2/PDX1.3 complex (intermediate) | | 分子名称: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | 著者 | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | 登録日 | 2018-10-17 | | 公開日 | 2019-04-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RYR
 
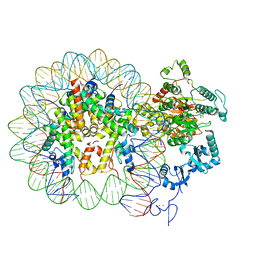 | | Nucleosome-CHD4 complex structure (single CHD4 copy) | | 分子名称: | Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | 著者 | Farnung, L, Ochmann, M, Cramer, P. | | 登録日 | 2019-06-11 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
8PKW
 
 | | Kelch domain of KEAP1 in complex with a ortho-dimethylbenzene linked cyclic peptide 5 (ortho-WRCDPETaEC). | | 分子名称: | (2-methylphenyl)methanol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Braun, M.B, Bischof, L, Hartmann, M.D. | | 登録日 | 2023-06-27 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Computational Prediction of Cyclic Peptide Structural Ensembles and Application to the Design of Keap1 Binders.
J.Chem.Inf.Model., 63, 2023
|
|
5DF5
 
 | | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution. | | 分子名称: | Cytochrome c, somatic, HEME C, ... | | 著者 | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | 登録日 | 2015-08-26 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.301 Å) | | 主引用文献 | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution.
To Be Published
|
|
8PKX
 
 | | Kelch domain of KEAP1 in complex with a ortho-dimethylbenzene linked cyclic peptide 11 (ortho-WRCNPETaEC). | | 分子名称: | (2-methylphenyl)methanol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Braun, M.B, Bischof, L, Hartmann, M.D. | | 登録日 | 2023-06-27 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Computational Prediction of Cyclic Peptide Structural Ensembles and Application to the Design of Keap1 Binders.
J.Chem.Inf.Model., 63, 2023
|
|
8PKV
 
 | | Kelch domain of KEAP1 in complex with a ortho-dimethylbenzene linked cyclic peptide 4 (ortho-WRCDEETGEC). | | 分子名称: | (2-methylphenyl)methanol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Braun, M.B, Bischof, L, Hartmann, M.D. | | 登録日 | 2023-06-27 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Computational Prediction of Cyclic Peptide Structural Ensembles and Application to the Design of Keap1 Binders.
J.Chem.Inf.Model., 63, 2023
|
|
