6YVK
 
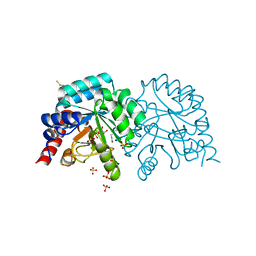 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 0.71 MGy exposure | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-04-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVL
 
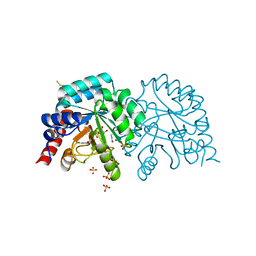 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 1.42 MGy exposure | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-04-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVM
 
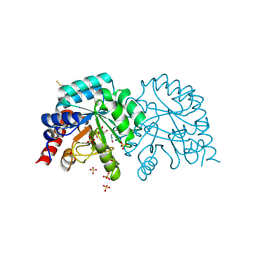 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.13 MGy exposure | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-04-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVN
 
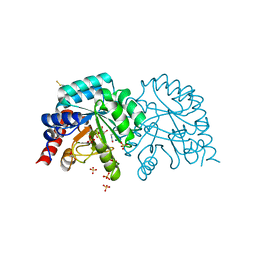 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.84 MGy exposure | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-04-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVO
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 3.55 MGy exposure | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-04-28 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
1W2Q
 
 | |
6YWT
 
 | |
6YWU
 
 | | Human OMPD-domain of UMPS (K314AcK) in complex with UMP at 1.1 Angstroms resolution | | 分子名称: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-04-30 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZWZ
 
 | |
6ZWY
 
 | |
6ZX1
 
 | | OMPD-domain of human UMPS in complex with 6-Aza-UMP at 1.0 Angstroms resolution | | 分子名称: | 6-AZA URIDINE 5'-MONOPHOSPHATE, PROLINE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-07-29 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZX3
 
 | | OMPD-domain of human UMPS in complex with 6-thiocarboxamido-UMP at 1.15 Angstroms resolution | | 分子名称: | GLYCEROL, PROLINE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Schimdt, T. | | 登録日 | 2020-07-29 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZX2
 
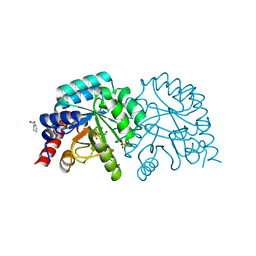 | | OMPD-domain of human UMPS in complex with 6-carboxamido-UMP at 1.2 Angstroms resolution | | 分子名称: | PROLINE, SULFATE ION, Uridine 5'-monophosphate synthase, ... | | 著者 | Tittmann, K, Rindfleisch, S, Schimdt, T. | | 登録日 | 2020-07-29 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZX0
 
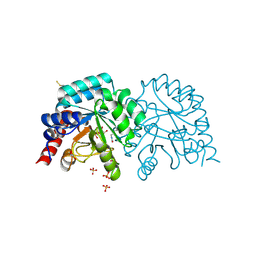 | | OMPD-domain of human UMPS in complex with the substrate OMP at 1.25 Angstroms resolution | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-07-29 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6P63
 
 | |
6NL2
 
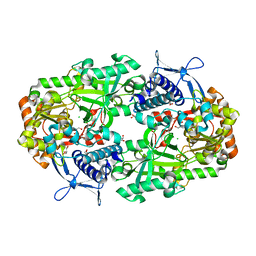 | | Apo NIS synthetase DesD variant R306Q | | 分子名称: | CHLORIDE ION, GLYCEROL, desferrioxamine E biosynthesis protein DesD | | 著者 | Hoffmann, K.M. | | 登録日 | 2019-01-07 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | A C-terminal Loop Mediates Cooperativity in the NIS Synthetase DesD
To Be Published
|
|
2KND
 
 | | Psb27 structure from Synechocystis | | 分子名称: | Photosystem II 11 kDa protein | | 著者 | Cormann, K.U, Nowaczyk, M.M, Bangert, J.-A, Ikeuchi, M, Roegner, M, Stoll, R. | | 登録日 | 2009-08-21 | | 公開日 | 2009-09-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of Psb27 in solution: implications for transient binding to photosystem II during biogenesis and repair
Biochemistry, 48, 2009
|
|
1RPM
 
 | |
7L06
 
 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to two copies of domain-swapped antibody 2G12 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | 著者 | Manne, K, Henderson, R, Acharya, P. | | 登録日 | 2020-12-11 | | 公開日 | 2020-12-30 | | 最終更新日 | 2021-06-09 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L6M
 
 | |
7L02
 
 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to one copy of domain-swapped antibody 2G12 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | 著者 | Manne, K, Henderson, R, Acharya, P. | | 登録日 | 2020-12-10 | | 公開日 | 2020-12-30 | | 最終更新日 | 2021-06-09 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L09
 
 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound domain-swapped antibody 2G12 from masked 3D refinement | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | 著者 | Manne, K, Henderson, R, Acharya, P. | | 登録日 | 2020-12-11 | | 公開日 | 2020-12-30 | | 最終更新日 | 2021-06-09 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L6O
 
 | | Cryo-EM structure of HIV-1 Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 - gp120, ... | | 著者 | Manne, K, Edwards, R.J, Acharya, P. | | 登録日 | 2020-12-23 | | 公開日 | 2021-04-14 | | 最終更新日 | 2021-06-09 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
5NIW
 
 | | Glucose oxydase mutant A2 | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Hoffmann, K, Frank, D. | | 登録日 | 2017-03-27 | | 公開日 | 2017-11-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Shuffling Active Site Substate Populations Affects Catalytic Activity: The Case of Glucose Oxidase.
ACS Catal, 7, 2017
|
|
8BVK
 
 | | The crystal structure of O-glycoside cleaving beta-eliminase from A. tumefaciens AtOGE | | 分子名称: | MANGANESE (II) ION, Xylose isomerase | | 著者 | Kuhlmann, K, Bitter, J, Pfeiffer, M, Nidetzky, B, Pavkov-Keller, T. | | 登録日 | 2022-12-04 | | 公開日 | 2023-11-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Enzymatic beta-elimination in natural product O- and C-glycoside deglycosylation.
Nat Commun, 14, 2023
|
|
