2FWE
 
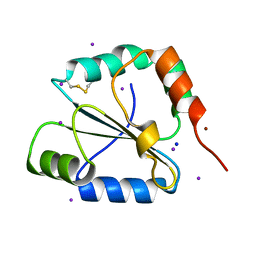 | | crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (oxidized form) | | Descriptor: | IODIDE ION, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
3M51
 
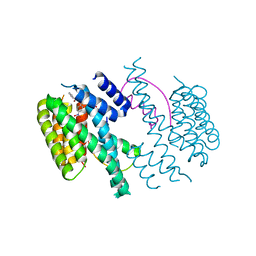 | | Structure of the 14-3-3/PMA2 complex stabilized by Pyrrolidone1 | | Descriptor: | 14-3-3-like protein C, 2-hydroxy-5-[(5S)-3-hydroxy-5-(4-nitrophenyl)-2-oxo-4-(phenylcarbonyl)-2,5-dihydro-1H-pyrrol-1-yl]benzoic acid, N.plumbaginifolia H+-translocating ATPase mRNA | | Authors: | Ottmann, C, Rose, R, Waldmann, H. | | Deposit date: | 2010-03-12 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Identification and structure of small-molecule stabilizers of 14-3-3 protein-protein interactions
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3M50
 
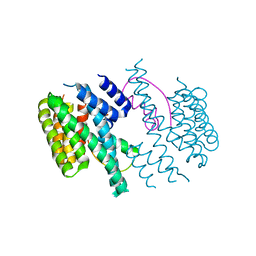 | | Structure of the 14-3-3/PMA2 complex stabilized by Epibestatin | | Descriptor: | 14-3-3-like protein C, N-[(2R,3R)-3-amino-2-hydroxy-4-phenylbutanoyl]-L-leucine, N.plumbaginifolia H+-translocating ATPase mRNA | | Authors: | Ottmann, C, Rose, R, Waldmann, H. | | Deposit date: | 2010-03-12 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification and structure of small-molecule stabilizers of 14-3-3 protein-protein interactions
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3DBS
 
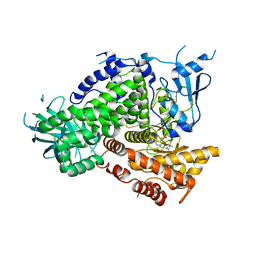 | | Structure of PI3K gamma in complex with GDC0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Wiesmann, C, Ultsch, M. | | Deposit date: | 2008-06-02 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer
J.Med.Chem., 51, 2008
|
|
1MWW
 
 | | THE STRUCTURE OF THE HYPOTHETICAL PROTEIN HI1388.1 FROM HAEMOPHILUS INFLUENZAE REVEALS A TAUTOMERASE/MIF FOLD | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, HYPOTHETICAL PROTEIN HI1388.1 | | Authors: | Lehmann, C, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-01 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of the Hypothetical Protein HI1388.1 from Haemophilus influenzae
To be Published
|
|
5HPA
 
 | |
2RKO
 
 | | Crystal Structure of the Vps4p-dimer | | Descriptor: | Vacuolar protein sorting-associated protein 4 | | Authors: | Hartmann, C, Gruetter, M.G. | | Deposit date: | 2007-10-17 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Vacuolar protein sorting: two different functional states of the AAA-ATPase Vps4p
J.Mol.Biol., 377, 2008
|
|
4RVK
 
 | | CHK1 kinase domain with diazacarbazole compound 8: N-[3-(6-cyano-9H-pyrrolo[2,3-b:5,4-c']dipyridin-3-yl)phenyl]acetamide | | Descriptor: | N-[3-(6-cyano-9H-pyrrolo[2,3-b:5,4-c']dipyridin-3-yl)phenyl]acetamide, Serine/threonine-protein kinase Chk1 | | Authors: | Wiesmann, C, Wu, P. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1.
J.Med.Chem., 58, 2015
|
|
4RVL
 
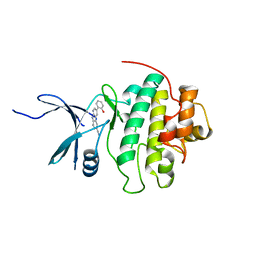 | | CHK1 kinase domain with diazacarbazole compound 7: 3-(2-hydroxyphenyl)-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile | | Descriptor: | 3-(2-hydroxyphenyl)-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Wiesmann, C, Wu, P. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1.
J.Med.Chem., 58, 2015
|
|
1FLT
 
 | | VEGF IN COMPLEX WITH DOMAIN 2 OF THE FLT-1 RECEPTOR | | Descriptor: | FMS-LIKE TYROSINE KINASE 1, VASCULAR ENDOTHELIAL GROWTH FACTOR | | Authors: | Wiesmann, C, De Vos, A.M. | | Deposit date: | 1997-11-20 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure at 1.7 A resolution of VEGF in complex with domain 2 of the Flt-1 receptor.
Cell(Cambridge,Mass.), 91, 1997
|
|
4QYH
 
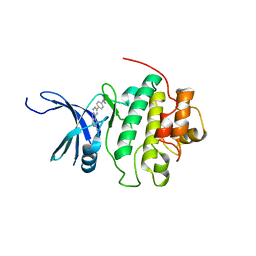 | | CHK1 kinase domain in complex with diazacarbazole GNE-783 | | Descriptor: | 3-[4-(4-methylpiperazin-1-yl)phenyl]-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Wiesmann, C, Wu, P. | | Deposit date: | 2014-07-24 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of the 1,7-diazacarbazole class of inhibitors of checkpoint kinase 1.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4QYG
 
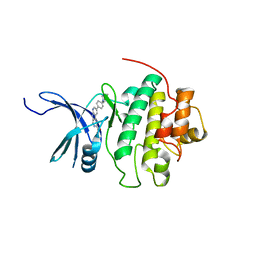 | | CHK1 kinase domain in complex with diazacarbazole compound 14 | | Descriptor: | 3-[4-(4-methylpiperazin-1-yl)phenyl]-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carboxylic acid, Serine/threonine-protein kinase Chk1 | | Authors: | Wiesmann, C, Wu, P. | | Deposit date: | 2014-07-24 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of the 1,7-diazacarbazole class of inhibitors of checkpoint kinase 1.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1HHZ
 
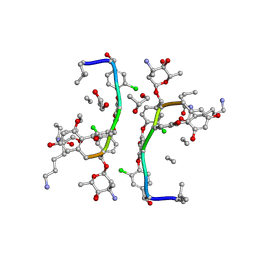 | | Deglucobalhimycin in complex with cell wall pentapeptide | | Descriptor: | (2R,4S,6S)-4-azanyl-4,6-dimethyl-oxane-2,5,5-triol, CELL WALL PEPTIDE, DEGLUCOBALHIMYCIN, ... | | Authors: | Lehmann, C, Bunkoczi, G, Sheldrick, G.M, Vertesy, L. | | Deposit date: | 2000-12-29 | | Release date: | 2003-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structures of Glycopeptide Antibiotics with Peptides that Model Bacterial Cell-Wall Precursors
J.Mol.Biol., 318, 2002
|
|
1GO6
 
 | | Balhimycin in complex with Lys-D-ala-D-ala | | Descriptor: | (2R,4S,6S)-4-azanyl-4,6-dimethyl-oxane-2,5,5-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Lehmann, C, Bunkoczi, G, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2001-10-19 | | Release date: | 2002-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structures of Glycopeptide Antibiotics with Peptides that Model Bacterial Cell-Wall Precursors
J.Mol.Biol., 318, 2002
|
|
1NNX
 
 | | Structure of the hypothetical protein ygiW from E. coli. | | Descriptor: | Protein ygiW, SULFATE ION | | Authors: | Lehmann, C, Galkin, A, Pullalarevu, S, Sarikaya, E, Krajewski, W, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the hypothetical protein ygiW from E. coli.
To be Published
|
|
1HH3
 
 | | Decaplanin first P21-Form | | Descriptor: | 4-epi-vancosamine, DECAPLANIN, GLYCEROL, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-19 | | Release date: | 2005-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
1HHF
 
 | | Decaplanin second P6122-Form | | Descriptor: | 4-epi-vancosamine, CHLORIDE ION, DECAPLANIN, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-22 | | Release date: | 2005-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
1HHU
 
 | | Balhimycin in complex with D-Ala-D-Ala | | Descriptor: | (2R,4S,6S)-4-azanyl-4,6-dimethyl-oxane-2,5,5-triol, (4S)-2-METHYL-2,4-PENTANEDIOL, BALHIMYCIN, ... | | Authors: | Lehmann, C, Bunkoczi, G, Sheldrick, G.M, Vertessy, L. | | Deposit date: | 2000-12-28 | | Release date: | 2003-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structures of Glycopeptide Antibiotics with Peptides that Model Bacterial Cell-Wall Precursors
J.Mol.Biol., 318, 2002
|
|
1HHC
 
 | | Crystal structure of Decaplanin - space group P21, second form | | Descriptor: | 4-epi-vancosamine, CITRIC ACID, DECAPLANIN, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-22 | | Release date: | 2005-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
1HHA
 
 | | Decaplanin first P6122-Form | | Descriptor: | 4-epi-vancosamine, DECAPLANIN, GLYCEROL, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-22 | | Release date: | 2005-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
6Y1W
 
 | | Xcc4156, a flavin-dependent halogenase from Xanthomonas campestris | | Descriptor: | (2S,3S)-butane-2,3-diol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Widmann, C, Ismail, M, Sewald, N, Niemann, H.H. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of apo flavin-dependent halogenase Xcc4156 hints at a reason for cofactor-soaking difficulties.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3BK9
 
 | | H55A mutant of tryptophan 2,3-dioxygenase from Xanthomonas campestris | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, Tryptophan 2,3-dioxygenase | | Authors: | Bruckmann, C, Mowat, C.G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling substrate binding
Biochemistry, 47, 2008
|
|
3O8I
 
 | | Structure of 14-3-3 isoform sigma in complex with a C-Raf1 peptide and a stabilizing small molecule fragment | | Descriptor: | 14-3-3 binding site peptide of RAF proto-oncogene serine/threonine-protein kinase, 14-3-3 protein sigma, 6,6-dihydroxy-1-methoxyhexan-2-one | | Authors: | Ottmann, C, Rose, R, Kaiser, M, Kuhenne, P. | | Deposit date: | 2010-08-03 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Impaired Binding of 14-3-3 to C-RAF in Noonan Syndrome Suggests New Approaches in Diseases with Increased Ras Signaling
Mol.Cell.Biol., 30, 2010
|
|
1HHY
 
 | | Deglucobalhimycin in complex with D-Ala-D-Ala | | Descriptor: | (2R,4S,6S)-4-azanyl-4,6-dimethyl-oxane-2,5,5-triol, D-ALANINE, DEGLUCOBALHIMYCIN, ... | | Authors: | Lehmann, C, Bunkoczi, G, Sheldrick, G.M, Vertesy, L. | | Deposit date: | 2000-12-29 | | Release date: | 2003-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structures of Glycopeptide Antibiotics with Peptides that Model Bacterial Cell-Wall Precursors
J.Mol.Biol., 318, 2002
|
|
1Z5Y
 
 | | Crystal Structure Of The Disulfide-Linked Complex Between The N-Terminal Domain Of The Electron Transfer Catalyst DsbD and The Cytochrome c Biogenesis Protein CcmG | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Thiol:disulfide interchange protein dsbD, ... | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Gruetter, M.G, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis and Kinetics of DsbD-Dependent Cytochrome c Maturation
STRUCTURE, 13, 2005
|
|
