1L1N
 
 | | POLIOVIRUS 3C PROTEINASE | | Descriptor: | Genome polyprotein: Picornain 3C | | Authors: | Mosimann, S.C, Chernaia, M.M, Sia, S, Plotch, S, James, M.N.G. | | Deposit date: | 2002-02-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined X-ray crystallographic structure of the poliovirus 3C gene product.
J.Mol.Biol., 273, 1997
|
|
4K7W
 
 | | Crystal structure of Zn3-hUb(human ubiquitin) adduct from a solution 100 mM zinc acetate/1.3 mM hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ZINC ION, ... | | Authors: | Fermani, S, Falini, G, Calvaresi, M, Bottoni, A, Arnesano, F, Natile, G. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Conformational selection of ubiquitin quaternary structures driven by zinc ions.
Chemistry, 19, 2013
|
|
1NHP
 
 | | CRYSTALLOGRAPHIC ANALYSES OF NADH PEROXIDASE CYS42ALA AND CYS42SER MUTANTS: ACTIVE SITE STRUCTURE, MECHANISTIC IMPLICATIONS, AND AN UNUSUAL ENVIRONMENT OF ARG303 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH PEROXIDASE, SULFATE ION | | Authors: | Mande, S.S, Claiborne, A, Hol, W.G.J. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analyses of NADH peroxidase Cys42Ala and Cys42Ser mutants: active site structures, mechanistic implications, and an unusual environment of Arg 303.
Biochemistry, 34, 1995
|
|
1KRS
 
 | |
7AAS
 
 | | Crystal structure of nitrosoglutathione reductase (GSNOR) from Chlamydomonas reinhardtii | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, S-(hydroxymethyl)glutathione dehydrogenase, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
4XXD
 
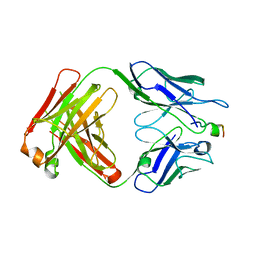 | | Crystal Structure of mid-region amyloid beta capture by solanezumab | | Descriptor: | Amyloid-beta fragment, Fab Heavy Chain, Fab Light Chain | | Authors: | Hermans, S.J, Crespi, G.A.N, Parker, M.W, Miles, L.A. | | Deposit date: | 2015-01-30 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular basis for mid-region amyloid-beta capture by leading Alzheimer's disease immunotherapies.
Sci Rep, 5, 2015
|
|
2PKQ
 
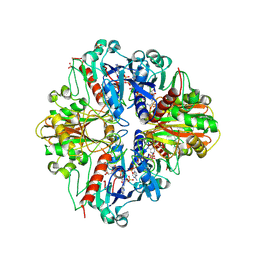 | | Crystal structure of the photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase, complexed with NADP | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, Glyceraldehyde-3-phosphate dehydrogenase B, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Fermani, S, Falini, G, Ripamonti, A. | | Deposit date: | 2007-04-18 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2G5X
 
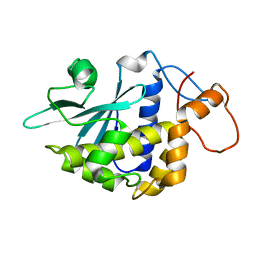 | | Crystal structure of lychnin a type 1 Ribosome Inactivating Protein (RIP) | | Descriptor: | Ribosome-inactivating protein | | Authors: | Fermani, S, Falini, G, Tosi, G, Ripamonti, A, Polito, L, Bolognesi, A, Stirpe, F. | | Deposit date: | 2006-02-23 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of lychnin a type 1 Ribosome Inactivating Protein (RIP)
To be Published
|
|
1J87
 
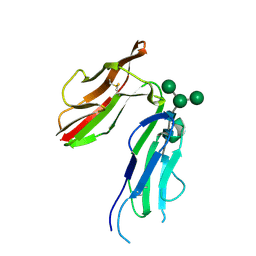 | | HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), HEXAGONAL CRYSTAL FORM 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, HIGH AFFINITY IMMUNOGLOBULIN EPSILON RECEPTOR ALPHA-SUBUNIT, ... | | Authors: | Garman, S.C, Sechi, S, Kinet, J.P, Jardetzky, T.S. | | Deposit date: | 2001-05-20 | | Release date: | 2001-08-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The analysis of the human high affinity IgE receptor Fc epsilon Ri alpha from multiple crystal forms.
J.Mol.Biol., 311, 2001
|
|
1J89
 
 | | HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), TETRAGONAL CRYSTAL FORM 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIGH AFFINITY IMMUNOGLOBULIN EPSILON RECEPTOR ALPHA-SUBUNIT, ... | | Authors: | Garman, S.C, Sechi, S, Kinet, J.P, Jardetzky, T.S. | | Deposit date: | 2001-05-20 | | Release date: | 2001-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | The analysis of the human high affinity IgE receptor Fc epsilon Ri alpha from multiple crystal forms.
J.Mol.Biol., 311, 2001
|
|
1J86
 
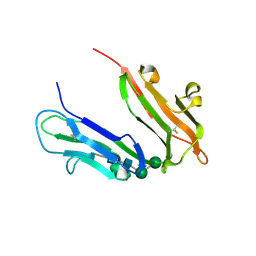 | | HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), MONOCLINIC CRYSTAL FORM 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garman, S.C, Sechi, S, Kinet, J.P, Jardetzky, T.S. | | Deposit date: | 2001-05-20 | | Release date: | 2001-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The analysis of the human high affinity IgE receptor Fc epsilon Ri alpha from multiple crystal forms.
J.Mol.Biol., 311, 2001
|
|
7AV7
 
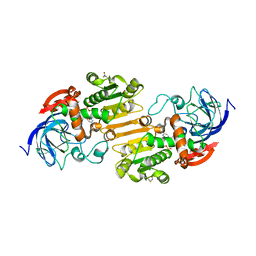 | | Crystal structure of S-nitrosylated nitrosoglutathione reductase(GSNOR)from Chlamydomonas reinhardtii, in complex with NAD+ | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-(hydroxymethyl)glutathione dehydrogenase, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
1J88
 
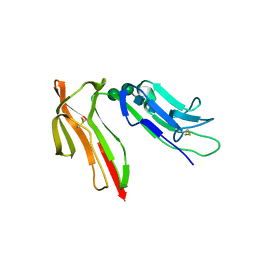 | | HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), TETRAGONAL CRYSTAL FORM 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIGH AFFINITY IMMUNOGLOBULIN EPSILON RECEPTOR ALPHA-SUBUNIT, ... | | Authors: | Garman, S.C, Sechi, S, Kinet, J.P, Jardetzky, T.S. | | Deposit date: | 2001-05-20 | | Release date: | 2001-08-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The analysis of the human high affinity IgE receptor Fc epsilon Ri alpha from multiple crystal forms.
J.Mol.Biol., 311, 2001
|
|
4XCL
 
 | |
2XQZ
 
 | | Neutron structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | BETA-LACTAMSE TOHO-1 | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
2X7W
 
 | | Crystal structure of Thermotoga maritima endonuclease IV in the presence of cadmium and zinc | | Descriptor: | BICINE, CADMIUM ION, PROBABLE ENDONUCLEASE 4, ... | | Authors: | Tomanicek, S.J, Hughes, R.C, Ng, J.D, Coates, L. | | Deposit date: | 2010-03-03 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure of the Endonuclease Iv Homologue from Thermotoga Maritima in the Presence of Active-Site Divalent Metal Ions
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1JMU
 
 | | Crystal Structure of the Reovirus mu1/sigma3 Complex | | Descriptor: | CHLORIDE ION, PROTEIN MU-1, SIGMA 3 PROTEIN, ... | | Authors: | Liemann, S, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2001-07-20 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reovirus membrane-penetration protein, Mu1, in a complex with is protector protein, Sigma3.
Cell(Cambridge,Mass.), 108, 2002
|
|
2XR0
 
 | | Room temperature X-ray structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | SULFATE ION, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
3N9B
 
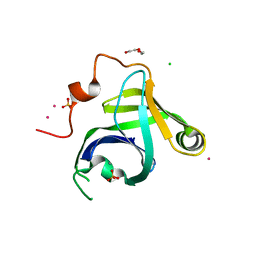 | | Crystal Structure of the P. aeruginosa LigD phosphoesterase domain | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Shuman, S, Nair, P, Smith, P. | | Deposit date: | 2010-05-28 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of bacterial LigD 3'-phosphoesterase unveils a DNA repair superfamily
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7AAU
 
 | | Crystal structure of nitrosoglutathione reductase from Chlamydomonas reinhardtii in complex with NAD+ | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
4Z0H
 
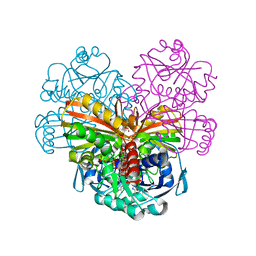 | | X-ray structure of cytoplasmic glyceraldehyde-3-phosphate dehydrogenase (GapC1) complexed with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase GAPC1, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Orru, R, Falini, G, Trost, P. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tuning Cysteine Reactivity and Sulfenic Acid Stability by Protein Microenvironment in Glyceraldehyde-3-Phosphate Dehydrogenases of Arabidopsis thaliana.
Antioxid. Redox Signal., 24, 2016
|
|
6ZGQ
 
 | | AceL NrdHF class 3 split intein GSH linked splice inactive variant - C124A, N146A | | Descriptor: | AceL NrdHF-1-1 Intein, IODIDE ION | | Authors: | Hoffmann, S, Mootz, H.D, Kuemmel, D, Singh, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Characterization of an Unusual and Naturally Split Class 3 Intein.
Chembiochem, 22, 2021
|
|
3N9D
 
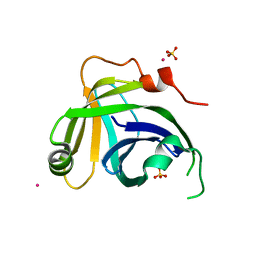 | | Monoclinic Structure of P. aeruginosa LigD phosphoesterase domain | | Descriptor: | MANGANESE (II) ION, Probable ATP-dependent DNA ligase, SULFATE ION, ... | | Authors: | Shuman, S, Nair, P, Smith, P. | | Deposit date: | 2010-05-28 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of bacterial LigD 3'-phosphoesterase unveils a DNA repair superfamily
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3RVD
 
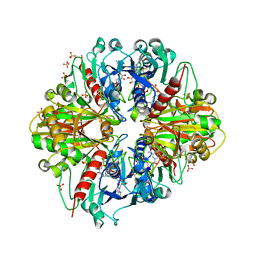 | | Crystal structure of the binary complex, obtained by soaking, of photosyntetic a4 glyceraldehyde 3-phosphate dehydrogenase (gapdh) with cp12-2, both from arabidopsis thaliana. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Thumiger, A, Falini, G, Marri, L, Sparla, F, Trost, P. | | Deposit date: | 2011-05-06 | | Release date: | 2012-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Selection and Folding-upon-binding of Intrinsically Disordered Protein CP12 Regulate Photosynthetic Enzymes Assembly.
J.Biol.Chem., 287, 2012
|
|
1JN0
 
 | | Crystal structure of the non-regulatory A4 isoform of spinach chloroplast glyceraldehyde-3-phosphate dehydrogenase complexed with NADP | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Fermani, S, Ripamonti, A, Sabatino, P, Zanotti, G, Scagliarini, S, Sparla, F, Trost, P, Pupillo, P. | | Deposit date: | 2001-07-21 | | Release date: | 2001-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the non-regulatory A(4 )isoform of spinach chloroplast glyceraldehyde-3-phosphate dehydrogenase complexed with NADP.
J.Mol.Biol., 314, 2001
|
|
