2XEL
 
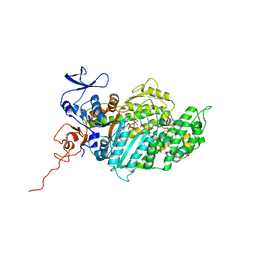 | | Molecular Mechanism of Pentachloropseudilin Mediated Inhibition of Myosin Motor Activity | | Descriptor: | 2,4-DICHLORO-6-(3,4,5-TRICHLORO-1H-PYRROL-2YL)PHENOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Chinthalapudi, K, Taft, M.H, Martin, R, Hartmann, F.K, Heissler, S.M, Tsiavaliaris, G, Gutzeit, H.O, Knoelker, H.J, Coluccio, L.M, Fedorov, R, Manstein, D.J. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism and Specificity of Pentachloropseudilin-Mediated Inhibition of Myosin Motor Activity.
J.Biol.Chem., 286, 2011
|
|
1UR5
 
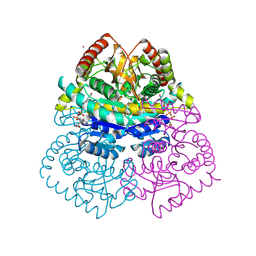 | | Stabilization of a Tetrameric Malate Dehydrogenase by Introduction of a Disulfide Bridge at the Dimer/Dimer Interface | | Descriptor: | CADMIUM ION, CHLORIDE ION, MALATE DEHYDROGENASE, ... | | Authors: | Bjork, A, Dalhus, B, Mantzilas, D, Eijsink, V.G.H, Sirevag, R. | | Deposit date: | 2003-10-27 | | Release date: | 2003-11-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stabilization of a Tetrameric Malate Dehydrogenase by Introduction of a Disulfide Bridge at the Dimer-Dimer Interface
J.Mol.Biol., 334, 2003
|
|
1UX5
 
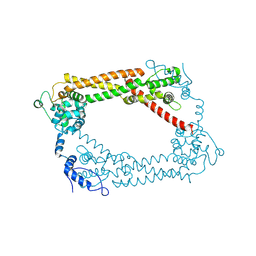 | | Crystal Structures of a Formin Homology-2 domain reveal a flexibly tethered dimer architecture | | Descriptor: | BNI1 PROTEIN | | Authors: | Xu, Y, Moseley, J.B, Sagot, I, Poy, F, Pellman, D, Goode, B.L, Eck, M.J. | | Deposit date: | 2004-02-19 | | Release date: | 2004-03-11 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of a Formin Homology-2 Domain Reveal a Tethered Dimer Architecture
Cell(Cambridge,Mass.), 116, 2004
|
|
1UXG
 
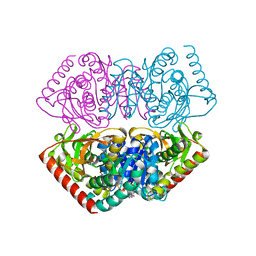 | | Large improvement in the thermal stability of a tetrameric malate dehydrogenase by single point mutations at the dimer-dimer interface. | | Descriptor: | FUMARIC ACID, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Bjork, A, Dalhus, B, Mantzilas, D, Eijsink, V.G.H, Sirevag, R. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Large Improvement in the Thermal Stability of a Tetrameric Malate Dehydrogenase by Single Point Mutations at the Dimer-Dimer Interface.
J.Mol.Biol., 341, 2004
|
|
2FLD
 
 | | I-MsoI Re-Designed for Altered DNA Cleavage Specificity | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*GP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*CP*TP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*CP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Ashworth, J, Duarte, C.M, Havranek, J.J, Sussman, D, Monnat, R.J, Stoddard, B.L, Baker, D. | | Deposit date: | 2006-01-05 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational redesign of endonuclease DNA binding and cleavage specificity.
Nature, 441, 2006
|
|
3RIU
 
 | |
1T61
 
 | | crystal structure of collagen IV NC1 domain from placenta basement membrane | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
1T4T
 
 | | arginase-dinor-NOHA complex | | Descriptor: | 3-{[(E)-AMINO(HYDROXYIMINO)METHYL]AMINO}ALANINE, Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Shoufa, H, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
1T60
 
 | | Crystal structure of Type IV collagen NC1 domain from bovine lens capsule | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
1UXI
 
 | | Large improvement in the thermal stability of a tetrameric malate dehydrogenase by single point mutations at the dimer-dimer interface | | Descriptor: | FUMARIC ACID, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bjork, A, Dalhus, B, Mantzilas, D, Eijsink, V.G.H, Sirevag, R. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Large Improvement in the Thermal Stability of a Tetrameric Malate Dehydrogenase by Single Point Mutations at the Dimer-Dimer Interface.
J.Mol.Biol., 341, 2004
|
|
1UXH
 
 | | Large improvement in the thermal stability of a tetrameric malate dehydrogenase by single point mutations at the dimer-dimer interface | | Descriptor: | FUMARIC ACID, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Bjork, A, Dalhus, B, Mantzilas, D, Eijsink, V.G.H, Sirevag, R. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Large Improvement in the Thermal Stability of a Tetrameric Malate Dehydrogenase by Single Point Mutations at the Dimer-Dimer Interface.
J.Mol.Biol., 341, 2004
|
|
1UPR
 
 | | Crystal structure of the PEPP1 pleckstrin homology domain in complex with Inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, PLECKSTRIN HOMOLOGY DOMAIN-CONTAINING FAMILY A MEMBER 4 | | Authors: | Milburn, C.C, Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Pleckstrin Homology Domain of Pepp1
To be Published
|
|
1UXJ
 
 | | Large improvement in the thermal stability of a tetrameric malate dehydrogenase by single point mutations at the dimer-dimer interface | | Descriptor: | CADMIUM ION, CHLORIDE ION, MALATE DEHYDROGENASE, ... | | Authors: | Bjork, A, Dalhus, B, Mantzilas, D, Eijsink, V.G.H, Sirevag, R. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Large Improvement in the Thermal Stability of a Tetrameric Malate Dehydrogenase by Single Point Mutations at the Dimer-Dimer Interface.
J.Mol.Biol., 341, 2004
|
|
1T9N
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, SULFATE ION, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-18 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis
by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
2HFW
 
 | | Structural and kinetic analysis of proton shuttle residues in the active site of human carbonic anhydrase III | | Descriptor: | Carbonic anhydrase 3, ZINC ION | | Authors: | Elder, I, Fisher, S.Z, Laipis, P.J, Tu, C.K, McKenna, R, Silverman, D.N. | | Deposit date: | 2006-06-26 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and kinetic analysis of proton shuttle residues in the active site of human carbonic anhydrase III.
Proteins, 68, 2007
|
|
1TS9
 
 | |
2EGL
 
 | | Crystal structure of Glu171 to Lys mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-01 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2EGS
 
 | | Crystal structure of Leu261 to Met mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-02 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
1M30
 
 | | Solution structure of N-terminal SH3 domain from oncogene protein c-Crk | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-26 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1M3C
 
 | | Solution structure of a circular form of the N-terminal SH3 domain (E132C, E133G, R191G mutant) from oncogene protein c-Crk | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1TE3
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | CHLORIDE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-24 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1TEQ
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, HYDROXIDE ION, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-25 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1M3B
 
 | | Solution structure of a circular form of the N-terminal SH3 domain (A134C, E135G, R191G mutant) from oncogene protein c-Crk. | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
2EGB
 
 | | Crystal structure of Glu140 to Asn mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-28 | | Release date: | 2007-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
3UNY
 
 | | Bacillus cereus phosphopentomutase T85E variant soaked with glucose 1,6-bisphosphate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
