4KWV
 
 | |
4KWW
 
 | | The crystal structure of human quinolinic acid phosphoribosyltransferase in complex with its inhibitor phthalic acid | | 分子名称: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], PHTHALIC ACID | | 著者 | Malik, S.S, Dimeka, P.N, Ncube, Z, Toth, E.A. | | 登録日 | 2013-05-24 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | The crystal structure of human quinolinic acid phosphoribosyltransferase in complex with its inhibitor phthalic acid.
Proteins, 82, 2014
|
|
2PRU
 
 | |
1JFI
 
 | | Crystal Structure of the NC2-TBP-DNA Ternary Complex | | 分子名称: | 5'-D(*G*GP*AP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*TP*CP*C)-3', TATA-BOX-BINDING PROTEIN (TBP), ... | | 著者 | Kamada, K, Shu, F, Chen, H, Malik, S, Stelzer, G, Roeder, R.G, Meisterernst, M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2001-06-20 | | 公開日 | 2001-07-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex.
Cell(Cambridge,Mass.), 106, 2001
|
|
5JXY
 
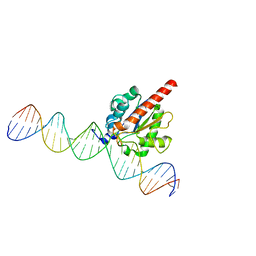 | | Enzyme-substrate complex of TDG catalytic domain bound to a G/U analog | | 分子名称: | DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | 著者 | Pidugu, L.S, Pozharski, E, Malik, S.S, Drohat, A.C. | | 登録日 | 2016-05-13 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
5HF7
 
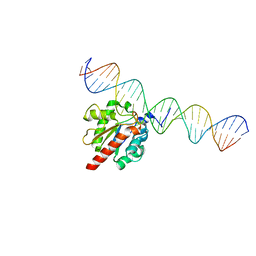 | | TDG enzyme-substrate complex | | 分子名称: | DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | 著者 | Pozharski, E, Malik, S.S, Drohat, A.C. | | 登録日 | 2016-01-06 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
5FF8
 
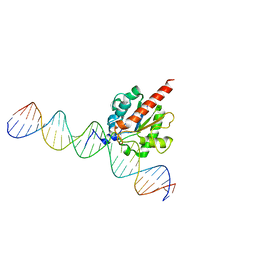 | | TDG enzyme-product complex | | 分子名称: | DNA, G/T mismatch-specific thymine DNA glycosylase | | 著者 | Pozharski, E, Malik, S.S, Drohat, A.C. | | 登録日 | 2015-12-18 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
4XEG
 
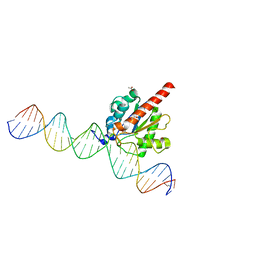 | | Structure of the enzyme-product complex resulting from TDG action on a G/hmU mismatch | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | 著者 | Pozharski, E, Malik, S.S, Drohat, A.C. | | 登録日 | 2014-12-23 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z47
 
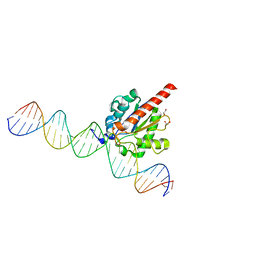 | | Structure of the enzyme-product complex resulting from TDG action on a GU mismatch in the presence of excess base | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, DNA, ... | | 著者 | Pozharski, E, Malik, S.S, Drohat, A.C. | | 登録日 | 2015-04-01 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z7Z
 
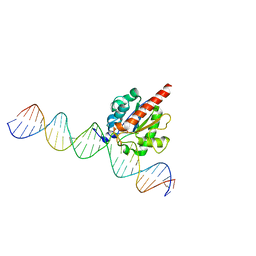 | | Structure of the enzyme-product complex resulting from TDG action on a GT mismatch in the presence of excess base | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | 著者 | Pozharski, E, Malik, S.S, Drohat, A.C. | | 登録日 | 2015-04-08 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z7B
 
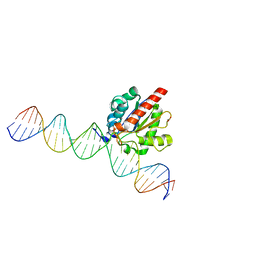 | | Structure of the enzyme-product complex resulting from TDG action on a GfC mismatch | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | 著者 | Pozharski, E, Malik, S.S, Drohat, A.C. | | 登録日 | 2015-04-07 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z3A
 
 | |
5CYS
 
 | |
