1MAP
 
 | |
1MAQ
 
 | |
1AKC
 
 | |
1AKA
 
 | |
1AKB
 
 | |
2IJQ
 
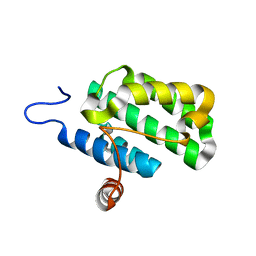 | | Crystal structure of protein rrnAC1037 from Haloarcula marismortui, Pfam DUF309 | | Descriptor: | Hypothetical protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the hypothetical Protein from Haloarcula marismortui
To be Published
|
|
2O34
 
 | | Crystal structure of protein DVU1097 from Desulfovibrio vulgaris Hildenborough, Pfam DUF375 | | Descriptor: | Hypothetical protein, SODIUM ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Desulfovibrio vulgaris Hildenborough
To be Published
|
|
2Q5U
 
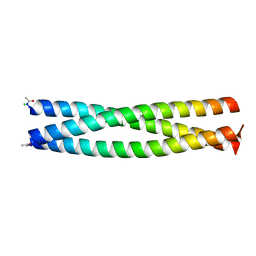 | | Crystal structure of IQN17 | | Descriptor: | CHLORIDE ION, Fusion protein between yeast variant GCN4 and HIVgp41 | | Authors: | Malashkevich, V.N, Eckert, D.M, Hong, L.H, Kim, P.S. | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibiting HIV Entry: Discovery of D-Peptide Inhibitors that Target the Gp41 Coiled-Coil Pocket
Cell(Cambridge,Mass.), 99, 1999
|
|
2Q7C
 
 | | Crystal structure of IQN17 | | Descriptor: | CHLORIDE ION, fusion protein between yeast variant GCN4 and HIVgp41 | | Authors: | Malashkevich, V.N, Eckert, D.M, Hong, L.H, Kim, P.S. | | Deposit date: | 2007-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibiting HIV Entry: Discovery of D-Peptide Inhibitors that Target the Gp41 Coiled-Coil Pocket
Cell(Cambridge,Mass.), 99, 1999
|
|
2Q3I
 
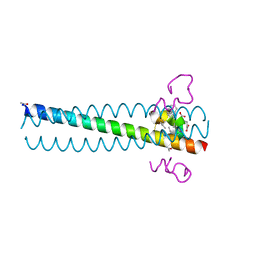 | | Crystal structure of the D10-P3/IQN17 complex: a D-peptide inhibitor of HIV-1 entry bound to the GP41 coiled-coil pocket | | Descriptor: | CHLORIDE ION, D-peptide, Fusion protein between the Coiled-Coil pocket of HIV GP41 and gcn4-PIQI | | Authors: | Malashkevich, V.N, Eckert, D.M, Hong, L.H, Carr, P.A, Kim, P.S. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibiting HIV Entry: Discovery of D-Peptide Inhibitors that Target the Gp41 Coiled-Coil Pocket
Cell(Cambridge,Mass.), 99, 1999
|
|
4MZZ
 
 | |
4N21
 
 | | Crystal structure of the GP2 Core Domain from the California Academy of Science Virus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GP2 Ectodomain | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Dai, Z, Toro, R, Lai, J.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Characterization of the Glycoprotein GP2 Core Domain from the CAS Virus, a Novel Arenavirus-Like Species.
J.Mol.Biol., 426, 2014
|
|
4NWS
 
 | |
4NSN
 
 | | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 030972, orthorhombic symmetry | | Descriptor: | ADENINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 030972, orthorhombic symmetry.
To be Published
|
|
4NS1
 
 | | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972 | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, GLYCEROL, Purine nucleoside phosphorylase, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972.
To be Published
|
|
4OXM
 
 | |
4P52
 
 | | Crystal structure of homoserine kinase from Cytophaga hutchinsonii ATCC 33406, NYSGRC Target 032717. | | Descriptor: | Homoserine kinase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-03-13 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of homoserine kinase from Cytophaga hutchinsonii ATCC 33406, NYSGRC Target 032717.
to be published
|
|
4PFQ
 
 | | Crystal structure of hypoxanthine phosphoribosyltransferase from Brachybacterium faecium DSM 4810, NYSGRC Target 029763. | | Descriptor: | Hypoxanthine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of hypoxanthine phosphoribosyltransferase from Brachybacterium faecium DSM 4810, NYSGRC Target 0299763.
to be published
|
|
4P67
 
 | |
4Q7U
 
 | |
4Q7T
 
 | |
4S1X
 
 | | Crystal structure of HA2-Del-L2seM, Central Coiled-Coil from Influenza Hemagglutinin HA2 without Heptad Repeat Stutter | | Descriptor: | GLYCEROL, Truncated hemagglutinin | | Authors: | Malashkevich, V.N, Higgins, C.D, Lai, J.R, Almo, S.C. | | Deposit date: | 2015-01-15 | | Release date: | 2015-04-01 | | Last modified: | 2015-06-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A switch from parallel to antiparallel strand orientation in a coiled-coil X-ray structure via two core hydrophobic mutations.
Biopolymers, 104, 2015
|
|
1AHG
 
 | | ASPARTATE AMINOTRANSFERASE HEXAMUTANT | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, TYROSINE | | Authors: | Malashkevich, V.N, Jansonius, J.N. | | Deposit date: | 1995-02-22 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alternating arginine-modulated substrate specificity in an engineered tyrosine aminotransferase.
Nat.Struct.Biol., 2, 1995
|
|
1AHY
 
 | | ASPARTATE AMINOTRANSFERASE HEXAMUTANT | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Malashkevich, V.N, Jansonius, J.N. | | Deposit date: | 1995-02-21 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alternating arginine-modulated substrate specificity in an engineered tyrosine aminotransferase.
Nat.Struct.Biol., 2, 1995
|
|
1AHF
 
 | | ASPARTATE AMINOTRANSFERASE HEXAMUTANT | | Descriptor: | ASPARTATE AMINOTRANSFERASE, INDOLYLPROPIONIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Malashkevich, V.N, Jansonius, J.N. | | Deposit date: | 1995-02-22 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alternating arginine-modulated substrate specificity in an engineered tyrosine aminotransferase.
Nat.Struct.Biol., 2, 1995
|
|
