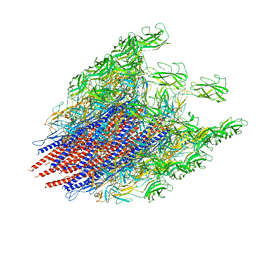
9IWQ
 
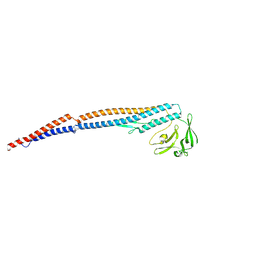 | | Salmonella enterica serovar Typhimurium FliC(G426A)delta(204-292) forming the L-type straight filament | | Descriptor: | Flagellin | | Authors: | Waraich, K, Makino, F, Miyata, T, Kinoshita, M, Minamino, T, Namba, K. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Salmonella enterica serovar Typhimurium FliC(G426A)delta(204-292) forming the L-type straight filament
To Be Published
|
|
6K9Q
 
 | | Structure of the native supercoiled hook as a universal joint | | Descriptor: | Flagellar hook protein FlgE | | Authors: | Kato, T, Miyata, T, Makino, F, Horvath, P, Namba, K. | | Deposit date: | 2019-06-17 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the native supercoiled flagellar hook as a universal joint.
Nat Commun, 10, 2019
|
|
4D3E
 
 | | Tetramer of IpaD, modified from 2J0O, fitted into negative stain electron microscopy reconstruction of the wild type tip complex from the type III secretion system of Shigella flexneri | | Descriptor: | INVASIN IPAD | | Authors: | Cheung, M, Shen, D.-K, Makino, F, Kato, T, Roehrich, D, Martinez-Argudo, I, Walker, M.L, Murillo, I, Liu, X, Pain, M, Brown, J, Frazer, G, Mantell, J, Mina, P, Todd, T, Sessions, R.B, Namba, K, Blocker, A.J. | | Deposit date: | 2014-10-21 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Three-Dimensional Electron Microscopy Reconstruction and Cysteine-Mediated Crosslinking Provide a Model of the T3Ss Needle Tip Complex.
Mol.Microbiol., 95, 2015
|
|
7BY0
 
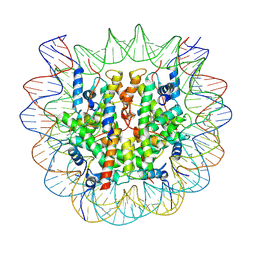 | | The cryo-EM structure of CENP-A nucleosome in complex with the phosphorylated CENP-C | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ariyoshi, M, Makino, F, Fukagawa, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C.
Embo J., 40, 2021
|
|
7BXT
 
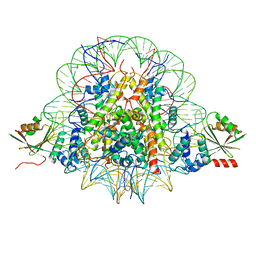 | | The cryo-EM structure of CENP-A nucleosome in complex with CENP-C peptide and CENP-N N-terminal domain | | Descriptor: | CENP-C, DNA (145-mer), Histone H2A type 1-B/E, ... | | Authors: | Ariyoshi, M, Makino, F, Fukagawa, T. | | Deposit date: | 2020-04-20 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C.
Embo J., 40, 2021
|
|
8W8M
 
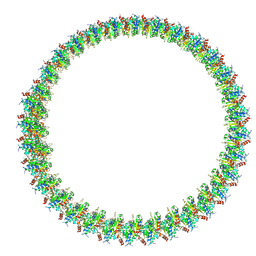 | | Cryo-EM structure of helical filament of MyD88 TIR | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Kasai, K, Imamura, K, Narita, A, Makino, F, Miyata, T, Kato, T, Namba, K, Onishi, H, Tochio, H. | | Deposit date: | 2023-09-04 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Signaling adopter protein in a self-assembled form
To Be Published
|
|
8XCN
 
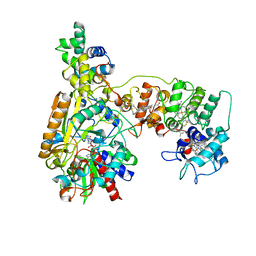 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1190A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
8XCM
 
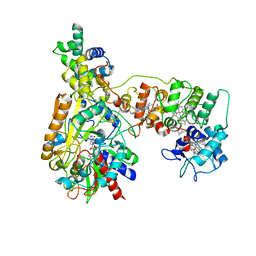 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146Q | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
8GY2
 
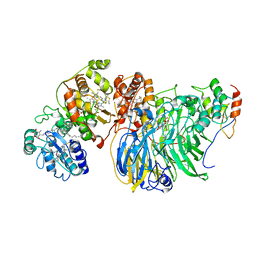 | | Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gluconobacter oxydans | | Descriptor: | Alcohol dehydrogenase (quinone), cytochrome c subunit, dehydrogenase subunit, ... | | Authors: | Adachi, T, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Experimental and Theoretical Insights into Bienzymatic Cascade for Mediatorless Bioelectrochemical Ethanol Oxidation with Alcohol and Aldehyde Dehydrogenases
Acs Catalysis, 13, 2023
|
|
8GY3
 
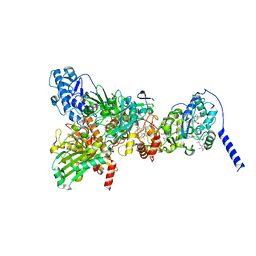 | | Cryo-EM Structure of Membrane-Bound Aldehyde Dehydrogenase from Gluconobacter oxydans | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), Cytochrome c subunit of aldehyde dehydrogenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Adachi, T, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Experimental and Theoretical Insights into Bienzymatic Cascade for Mediatorless Bioelectrochemical Ethanol Oxidation with Alcohol and Aldehyde Dehydrogenases
Acs Catalysis, 13, 2023
|
|
8JEK
 
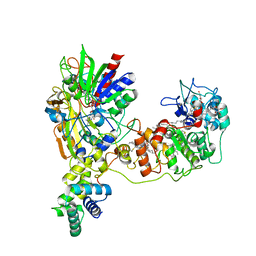 | | Cryo-EM Structure of K-ferricyanide Oxidized Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry.
Acs Catalysis, 13, 2023
|
|
8JEJ
 
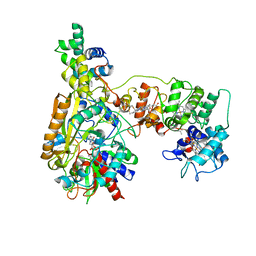 | | Cryo-EM Structure of Na-dithionite Reduced Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry.
Acs Catalysis, 13, 2023
|
|
8K6J
 
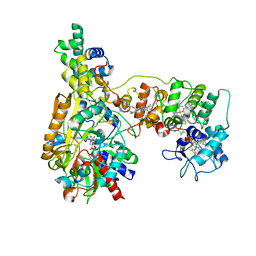 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-H1147A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase (H1147A) large subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-07-25 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
8K6K
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase (N1146A) large subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-07-25 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
7CLR
 
 | | CryoEM structure of S.typhimurium flagellar LP ring | | Descriptor: | Flagellar L-ring protein, Flagellar P-ring protein | | Authors: | Yamaguchi, T, Makino, F, Miyata, T, Minamino, T, Kato, T, Namba, K. | | Deposit date: | 2020-07-21 | | Release date: | 2021-06-02 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the molecular bushing of the bacterial flagellar motor.
Nat Commun, 12, 2021
|
|
7D84
 
 | | 34-fold symmetry Salmonella S ring formed by full-length FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Kawamoto, A, Miyata, T, Makino, F, Kinoshita, M, Minamino, T, Imada, K, Kato, T, Namba, K. | | Deposit date: | 2020-10-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries.
Nat Commun, 12, 2021
|
|
7VW6
 
 | | Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yoshikawa, T, Makino, F, Miyata, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Multiple electron transfer pathways of tungsten-containing formate dehydrogenase in direct electron transfer-type bioelectrocatalysis.
Chem.Commun.(Camb.), 58, 2022
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7W2J
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
7WSQ
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
7Y7I
 
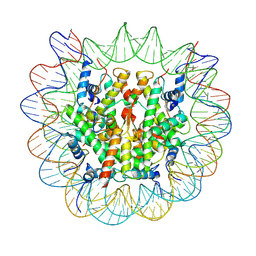 | | chicken KNL2 in complex with the CENP-A nucleosome | | Descriptor: | Chains: I, Chains: J, Histone H2A type 1-B/E, ... | | Authors: | Ariyoshi, M, Jiang, H, Makino, F, Fukagawa, T. | | Deposit date: | 2022-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The cryo-EM structure of the CENP-A nucleosome in complex with ggKNL2.
Biorxiv, 2022
|
|
