1N51
 
 | | Aminopeptidase P in complex with the inhibitor apstatin | | Descriptor: | MANGANESE (II) ION, Xaa-Pro aminopeptidase, apstatin | | Authors: | Graham, S.C, Maher, M.J, Lee, M.H, Simmons, W.H, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-11-03 | | Release date: | 2003-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OV8
 
 | | Auracyanin B structure in space group, P65 | | Descriptor: | Auracyanin B, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Lee, M, Maher, M.J, Freeman, H.C, Guss, J.M. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Auracyanin B structure in space group P6(5).
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2C9R
 
 | | apo-H91F CopC | | Descriptor: | COPPER RESISTANCE PROTEIN C, SODIUM ION | | Authors: | Zhang, L, Koay, M, Maher, M.J, Xiao, Z, Wedd, A.G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intermolecular Transfer of Copper Ions from the Copc Protein of Pseudomonas Syringae. Crystal Structures of Fully Loaded Cu(I)Cu(II) Forms.
J.Am.Chem.Soc., 128, 2006
|
|
2C9P
 
 | | Cu(I)Cu(II)-CopC at pH 4.5 | | Descriptor: | COPPER (II) ION, COPPER RESISTANCE PROTEIN C, NITRATE ION | | Authors: | Zhang, L, Koay, M, Maher, M.J, Xiao, Z, Wedd, A.G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Intermolecular Transfer of Copper Ions from the Copc Protein of Pseudomonas Syringae. Crystal Structures of Fully Loaded Cu(I)Cu(II) Forms.
J.Am.Chem.Soc., 128, 2006
|
|
2EG6
 
 | |
2EG8
 
 | | The crystal structure of E. coli dihydroorotase complexed with 5-fluoroorotic acid | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-02-28 | | Release date: | 2007-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Ligand-free and Inhibitor Complexes of Dihydroorotase from Escherichia coli: Implications for Loop Movement in Inhibitor Design
J.Mol.Biol., 370, 2007
|
|
2E25
 
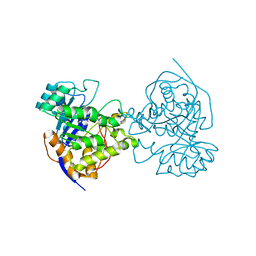 | | The Crystal Structure of the T109S mutant of E. coli Dihydroorotase complexed with an inhibitor 5-fluoroorotate | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2006-11-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the T109S mutant of Escherichia coli dihydroorotase complexed with the inhibitor 5-fluoroorotate: catalytic activity is reflected by the crystal form
Acta Crystallogr.,Sect.F, 63, 2007
|
|
4GAV
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with quinone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase, UBIQUINONE-2 | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G9K
 
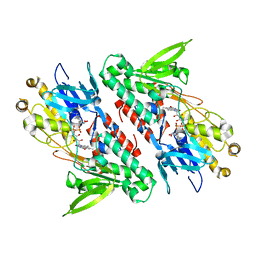 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GAP
 
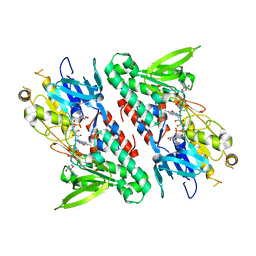 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with NAD+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3LX5
 
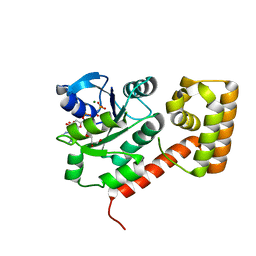 | | Crystal structure of mGMPPNP-bound NFeoB from S. thermophilus | | Descriptor: | 2-amino-9-(5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]-3-O-{[2-(methylamino)phenyl]carbonyl}-beta-D-erythro-pentofuranosyl-2-ulose)-1,9-dihydro-6H-purin-6-one, Ferrous iron uptake transporter protein B, GLYCEROL, ... | | Authors: | Ash, M.R, Guilfoyle, A, Maher, M.J, Clarke, R.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potassium-activated GTPase reaction in the G Protein-coupled ferrous iron transporter B.
J.Biol.Chem., 285, 2010
|
|
3SS8
 
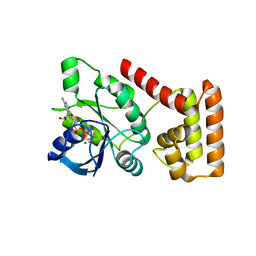 | | Crystal structure of NFeoB from S. thermophilus bound to GDP.AlF4- and K+ | | Descriptor: | Ferrous iron uptake transporter protein B, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The initiation of GTP hydrolysis by the G-domain of FeoB: insights from a transition-state complex structure
Plos One, 6, 2011
|
|
3LX8
 
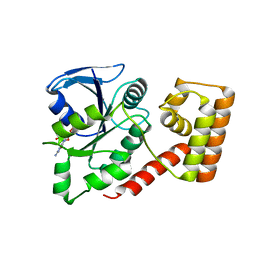 | | Crystal structure of GDP-bound NFeoB from S. thermophilus | | Descriptor: | Ferrous iron uptake transporter protein B, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ash, M.R, Guilfoyle, A, Maher, M.J, Clarke, R.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potassium-activated GTPase reaction in the G Protein-coupled ferrous iron transporter B.
J.Biol.Chem., 285, 2010
|
|
3TU6
 
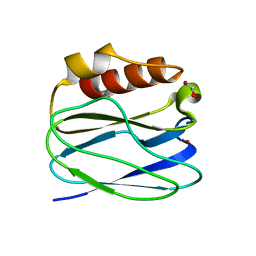 | | The Structure of a Pseudoazurin From Sinorhizobium meliltoi | | Descriptor: | COPPER (II) ION, GLYCEROL, Pseudoazurin (Blue copper protein) | | Authors: | Laming, E.M, McGrath, A.P, Guss, J.M, Maher, M.J. | | Deposit date: | 2011-09-16 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray crystal structure of a pseudoazurin from Sinorhizobium meliloti.
J.Inorg.Biochem., 115, 2012
|
|
4PW9
 
 | |
3TAH
 
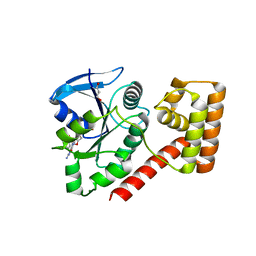 | | Crystal structure of an S. thermophilus NFeoB N11A mutant bound to mGDP | | Descriptor: | 3'-O-(N-methylanthraniloyl)guanosine-5'-diphosphate, Ferrous iron uptake transporter protein B, GLYCEROL, ... | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-08-04 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of an N11A mutant of the G-protein domain of FeoB
Acta Crystallogr.,Sect.F, 67, 2011
|
|
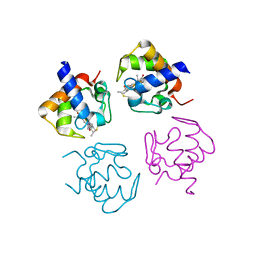
4PWA
 
 | |
4PW3
 
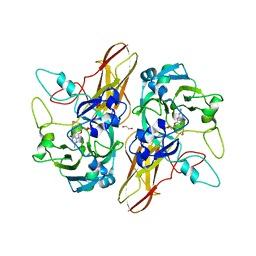 | |
1QHQ
 
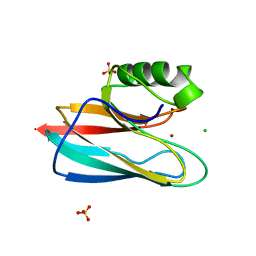 | | AURACYANIN, A BLUE COPPER PROTEIN FROM THE GREEN THERMOPHILIC PHOTOSYNTHETIC BACTERIUM CHLOROFLEXUS AURANTIACUS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (AURACYANIN), ... | | Authors: | Bond, C.S, Blankenship, R.E, Freeman, H.C, Guss, J.M, Maher, M, Selvaraj, F, Wilce, M.C.J, Willingham, K. | | Deposit date: | 1999-05-25 | | Release date: | 2001-03-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of auracyanin, a "blue" copper protein from the green thermophilic photosynthetic bacterium Chloroflexus aurantiacus.
J.Mol.Biol., 306, 2001
|
|
3MJM
 
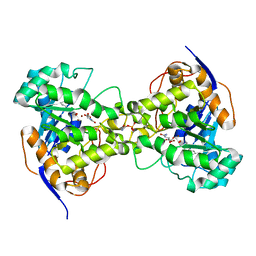 | | His257Ala mutant of dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Ernberg, K.E, Guss, J.M, Lee, M, Maher, M.J. | | Deposit date: | 2010-04-13 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | His257Ala mutant of dihydroorotase from E. coli
To be Published
|
|
3N7E
 
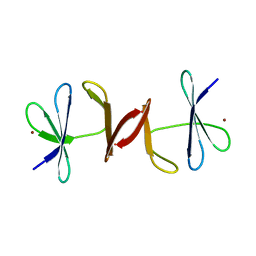 | | Crystal structure of CopK bound to Cu(II) | | Descriptor: | COPPER (II) ION, Copper resistance protein K | | Authors: | Ash, M.-R, Maher, M.J. | | Deposit date: | 2010-05-27 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Two new crystal forms of copper resistance protein CopK
To be Published
|
|
3N7D
 
 | |
1RUT
 
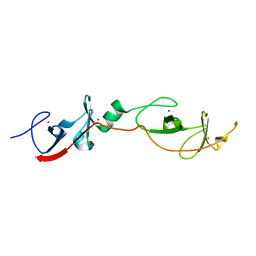 | | Complex of LMO4 LIM domains 1 and 2 with the ldb1 LID domain | | Descriptor: | Fusion protein of Lmo4 protein and LIM domain-binding protein 1, ZINC ION | | Authors: | Deane, J.E, Ryan, D.P, Maher, M.J, Kwan, A.H.Y, Bacca, M, Mackay, J.P, Guss, J.M, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2003-12-11 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tandem LIM domains provide synergistic binding in the LMO4:Ldb1 complex
Embo J., 23, 2004
|
|
6X9H
 
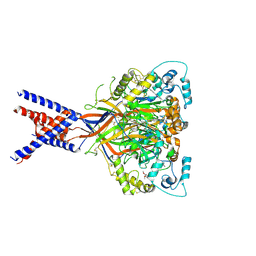 | | Molecular mechanism and structural basis of small-molecule modulation of acid-sensing ion channel 1 (ASIC1) | | Descriptor: | 2-[4-(3,4-dimethoxyphenoxy)phenyl]-1H-benzimidazole-6-carboximidamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, ... | | Authors: | Liu, Y, Ma, J, DesJarlais, R.L, Hagan, R, Rech, J, Lin, D, Liu, C, Miller, R, Schoellerman, J, Luo, J, Letavic, M, Grasberger, B, Maher, M. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Molecular mechanism and structural basis of small-molecule modulation of the gating of acid-sensing ion channel 1.
Commun Biol, 4, 2021
|
|
5VDE
 
 | |
