3K2P
 
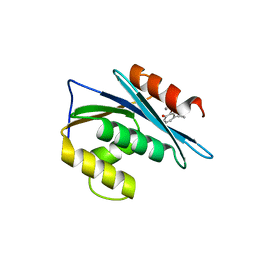 | | HIV-1 Reverse Transcriptase Isolated RnaseH Domain with the Inhibitor beta-thujaplicinol Bound at the Active Site | | Descriptor: | 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, MANGANESE (II) ION, Reverse Transcriptase | | Authors: | Pauly, T.A, Himmel, D.M, Maegley, K, Arnold, E. | | Deposit date: | 2009-09-30 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site.
Structure, 17, 2009
|
|
1EFY
 
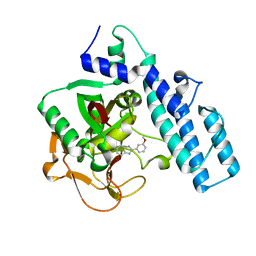 | | CRYSTAL STRUCTURE OF THE CATALYTIC FRAGMENT OF POLY (ADP-RIBOSE) POLYMERASE COMPLEXED WITH A BENZIMIDAZOLE INHIBITOR | | Descriptor: | 2-(3'-METHOXYPHENYL) BENZIMIDAZOLE-4-CARBOXAMIDE, POLY (ADP-RIBOSE) POLYMERASE | | Authors: | White, A.W, Almassy, R, Calvert, A.H, Curtin, N.J, Griffin, R.J, Hostomsky, Z, Maegley, K, Newell, D.R, Srinivasan, S, Golding, B.T. | | Deposit date: | 2000-02-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance-modifying agents. 9. Synthesis and biological properties of benzimidazole inhibitors of the DNA repair enzyme poly(ADP-ribose) polymerase.
J.Med.Chem., 43, 2000
|
|
3IG1
 
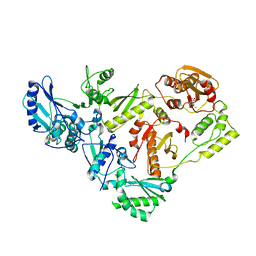 | | HIV-1 Reverse Transcriptase with the Inhibitor beta-Thujaplicinol Bound at the RNase H Active Site | | Descriptor: | 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, HIV-1 Reverse Transcriptase p51 subunit, HIV-1 Reverse Transcriptase p66 subunit, ... | | Authors: | Himmel, D.M, Maegley, K.A, Pauly, T.A, Arnold, E. | | Deposit date: | 2009-07-27 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site.
Structure, 17, 2009
|
|
1XR6
 
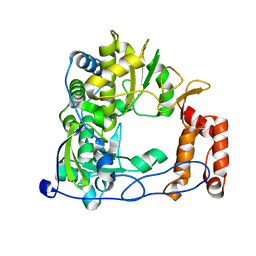 | | Crystal Structure of RNA-dependent RNA Polymerase 3D from human rhinovirus serotype 1B | | Descriptor: | Genome polyprotein, POTASSIUM ION | | Authors: | Love, R.A, Maegley, K.A, Yu, X, Ferre, R.A, Lingardo, L.K, Diehl, W, Parge, H.E, Dragovich, P.S, Fuhrman, S.A. | | Deposit date: | 2004-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of the RNA-Dependent RNA Polymerase from Human Rhinovirus: A Dual-Function Target for Common Cold Antiviral Therapy
Structure, 12, 2004
|
|
1XR5
 
 | | Crystal Structure of the RNA-dependent RNA Polymerase 3D from human rhinovirus serotype 14 | | Descriptor: | Genome polyprotein, SAMARIUM (III) ION | | Authors: | Love, R.A, Maegley, K.A, Yu, X, Ferre, R.A, Lingardo, L.K, Diehl, W, Parge, H.E, Dragovich, P.S, Fuhrman, S.A. | | Deposit date: | 2004-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the RNA-Dependent RNA Polymerase from Human Rhinovirus: A Dual-Function Target for Common Cold Antiviral Therapy
Structure, 12, 2004
|
|
1XR7
 
 | | Crystal structure of RNA-dependent RNA Polymerase 3D from human rhinovirus serotype 16 | | Descriptor: | Genome polyprotein | | Authors: | Love, R.A, Maegley, K.A, Yu, X, Ferre, R.A, Lingardo, L.K, Diehl, W, Parge, H.E, Dragovich, P.S, Fuhrman, S.A. | | Deposit date: | 2004-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the RNA-Dependent RNA Polymerase from Human Rhinovirus: A Dual-Function Target for Common Cold Antiviral Therapy
Structure, 12, 2004
|
|
3RLQ
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5- carbonitrile | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RLP
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RLR
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
