2IWT
 
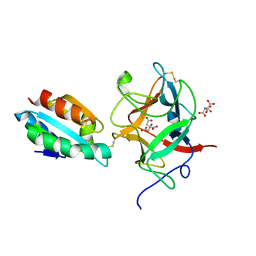 | | Thioredoxin h2 (HvTrxh2) in a mixed disulfide complex with the target protein BASI | | Descriptor: | ALPHA-AMYLASE/SUBTILISIN INHIBITOR, CITRATE ANION, THIOREDOXIN H ISOFORM 2 | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2006-07-04 | | Release date: | 2006-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Target Protein Recognition by the Protein Disulfide Reductase Thioredoxin.
Structure, 14, 2006
|
|
4B2Z
 
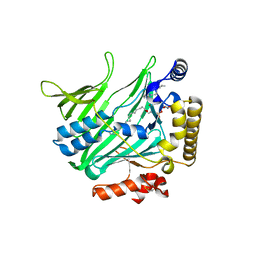 | | Structure of Osh6 in complex with phosphatidylserine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Maeda, K, Anand, K, Chiapparino, A, Kumar, A, Poletto, M, Kaksonen, M, Gavin, A.C. | | Deposit date: | 2012-07-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interactome Map Uncovers Phosphatidylserine Transport by Oxysterol-Binding Proteins
Nature, 501, 2013
|
|
2VLT
 
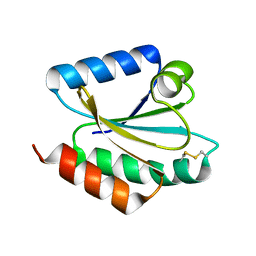 | | Crystal structure of barley thioredoxin h isoform 2 in the oxidized state | | Descriptor: | THIOREDOXIN H ISOFORM 2. | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2008-01-16 | | Release date: | 2008-04-29 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Barley Thioredoxin H Isoforms Hvtrxh1 and Hvtrxh2 Reveal Features Involved in Protein Recognition and Possibly in Discriminating the Isoform Specificity.
Protein Sci., 17, 2008
|
|
2VLV
 
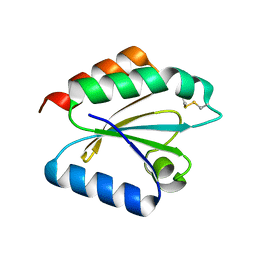 | | Crystal structure of barley thioredoxin h isoform 2 in partially radiation-reduced state | | Descriptor: | THIOREDOXIN H ISOFORM 2. | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2008-01-16 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Barley Thioredoxin H Isoforms Hvtrxh1 and Hvtrxh2 Reveal Features Involved in Protein Recognition and Possibly in Discriminating the Isoform Specificity.
Protein Sci., 17, 2008
|
|
2VM1
 
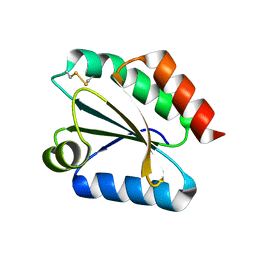 | | Crystal structure of barley thioredoxin h isoform 1 crystallized using ammonium sulfate as precipitant | | Descriptor: | SULFATE ION, THIOREDOXIN H ISOFORM 1. | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2008-01-21 | | Release date: | 2008-04-29 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Barley Thioredoxin H Isoforms Hvtrxh1 and Hvtrxh2 Reveal Features Involved in Protein Recognition and Possibly in Discriminating the Isoform Specificity.
Protein Sci., 17, 2008
|
|
2VLU
 
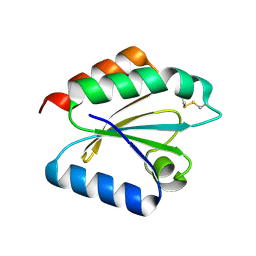 | | Crystal structure of barley thioredoxin h isoform 2 in partially radiation-reduced state | | Descriptor: | THIOREDOXIN H ISOFORM 2. | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2008-01-16 | | Release date: | 2008-04-29 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Barley Thioredoxin H Isoforms Hvtrxh1 and Hvtrxh2 Reveal Features Involved in Protein Recognition and Possibly in Discriminating the Isoform Specificity.
Protein Sci., 17, 2008
|
|
2VM2
 
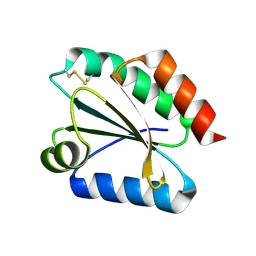 | | Crystal structure of barley thioredoxin h isoform 1 crystallized using PEG as precipitant | | Descriptor: | THIOREDOXIN H ISOFORM 1. | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2008-01-21 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Barley Thioredoxin H Isoforms Hvtrxh1 and Hvtrxh2 Reveal Features Involved in Protein Recognition and Possibly in Discriminating the Isoform Specificity.
Protein Sci., 17, 2008
|
|
1J1E
 
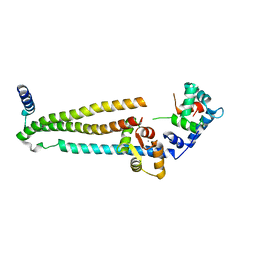 | | Crystal structure of the 52kDa domain of human cardiac troponin in the Ca2+ saturated form | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Takeda, S, Yamashita, A, Maeda, K, Maeda, Y. | | Deposit date: | 2002-12-03 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the core domain of human cardiac troponin in the Ca2+-saturated form
Nature, 424, 2003
|
|
1J1D
 
 | | Crystal structure of the 46kDa domain of human cardiac troponin in the Ca2+ saturated form | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Takeda, S, Yamashita, A, Maeda, K, Maeda, Y. | | Deposit date: | 2002-12-03 | | Release date: | 2003-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of the core domain of human cardiac troponin in the Ca2+-saturated form
Nature, 424, 2003
|
|
1A2X
 
 | | COMPLEX OF TROPONIN C WITH A 47 RESIDUE (1-47) FRAGMENT OF TROPONIN I | | Descriptor: | CALCIUM ION, TROPONIN C, TROPONIN I | | Authors: | Vassylyev, D.G, Takeda, S, Wakatsuki, S, Maeda, K, Maeda, Y. | | Deposit date: | 1998-01-13 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of troponin C in complex with troponin I fragment at 2.3-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1IZN
 
 | |
2EFS
 
 | | Crystal structure of the C-terminal tropomyosin fragment with N- and C-terminal extensions of the leucine zipper at 2.0 angstroms resolution | | Descriptor: | General control protein GCN4 and Tropomyosin 1 alpha chain | | Authors: | Minakata, S, Nitanai, Y, Maeda, K, Oda, N, Wakabayashi, K, Maeda, Y. | | Deposit date: | 2007-02-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two crystal structures of tropomyosin C-terminal fragment 176-273: exposure of the hydrophobic core to the solvent destabilizes the tropomyosin molecule
To be Published
|
|
2EFR
 
 | | Crystal structure of the c-terminal tropomyosin fragment with N- and C-terminal extensions of the leucine zipper at 1.8 angstroms resolution | | Descriptor: | General control protein GCN4 and Tropomyosin 1 alpha chain | | Authors: | Minakata, S, Nitanai, Y, Maeda, K, Oda, N, Wakabayashi, K, Maeda, Y. | | Deposit date: | 2007-02-23 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two crystal structures of tropomyosin C-terminal fragment 176-273: exposure of the hydrophobic core to the solvent destabilizes the tropomyosin molecule
To be Published
|
|
2D3E
 
 | |
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
2ZGY
 
 | | PARM with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid segregation protein parM | | Authors: | Popp, D, Narita, A, Oda, T, Fujisawa, T, Matsuo, H, Nitanai, Y, Iwasa, M, Maeda, K, Onishi, H, Maeda, Y. | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability
Embo J., 27, 2008
|
|
2ZGZ
 
 | | PARM with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plasmid segregation protein parM | | Authors: | Popp, D, Narita, A, Oda, T, Fujisawa, T, Matsuo, H, Nitanai, Y, Iwasa, M, Maeda, K, Onishi, H, Maeda, Y. | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability
Embo J., 27, 2008
|
|
4A6F
 
 | | Crystal structure of Slm1-PH domain in complex with Phosphoserine | | Descriptor: | PHOSPHATE ION, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE-BINDING PROTEIN SLM1, PHOSPHOSERINE | | Authors: | Anand, K, Maeda, K, Gavin, A.C. | | Deposit date: | 2011-11-02 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Analyses of Slm1-Ph Domain Demonstrate Ligand Binding in the Non-Canonical Site
Plos One, 7, 2012
|
|
4A6K
 
 | | Crystal structure of Slm1-PH domain in complex with D-myo-Inositol-4- phosphate | | Descriptor: | D-MYO-INOSITOL-4-PHOSPHATE, PHOSPHATE ION, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE-BINDING PROTEIN SLM1 | | Authors: | Anand, K, Maeda, K, Gavin, A.C. | | Deposit date: | 2011-11-04 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analyses of Slm1-Ph Domain Demonstrate Ligand Binding in the Non-Canonical Site
Plos One, 7, 2012
|
|
4A6H
 
 | | Crystal structure of Slm1-PH domain in complex with Inositol-4- phosphate | | Descriptor: | D-MYO-INOSITOL-4-PHOSPHATE, PHOSPHATE ION, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE-BINDING PROTEIN SLM1 | | Authors: | Anand, K, Maeda, K, Gavin, A.C. | | Deposit date: | 2011-11-03 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural Analyses of Slm1-Ph Domain Demonstrate Ligand Binding in the Non-Canonical Site
Plos One, 7, 2012
|
|
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | Authors: | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
2ZHC
 
 | | ParM filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid segregation protein parM | | Authors: | Popp, D, Narita, A, Oda, T, Fujisawa, T, Matsuo, H, Nitanai, Y, Iwasa, M, Maeda, K, Onishi, H, Maeda, Y. | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability
Embo J., 27, 2008
|
|
