4UXS
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4UZ9
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM - CRYSTAL FORM VII - SOS COMPLEX - 2.2A | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Notum Deacylates Wnt Proteins to Suppress Signalling Activity.
Nature, 519, 2015
|
|
4UZL
 
 | |
7QKE
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with inhibitor (surface entropy reduction mutant) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid C26-monooxygenase, ethyl 1-(cyclohexylmethyl)-5-pyridin-4-yl-indole-2-carboxylate | | Authors: | Snee, M, Tunnicliffe, R, Leys, D, Levy, C, Katariya, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
4V5A
 
 | | Structure of the Ribosome Recycling Factor bound to the Thermus thermophilus 70S ribosome with mRNA, ASL-Phe and tRNA-fMet | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Petry, S, Dunham, C.M, Selmer, M, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2007-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the ribosome recycling factor bound to the ribosome.
Nat. Struct. Mol. Biol., 14, 2007
|
|
4V4J
 
 | | Interactions and Dynamics of the Shine-Dalgarno Helix in the 70S Ribosome. | | Descriptor: | 16S RNA, 23S LARGE SUBUNIT RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Trakhanov, S, Asahara, H, Laurberg, M, Noller, H.F. | | Deposit date: | 2007-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.83 Å) | | Cite: | Interactions and dynamics of the Shine Dalgarno helix in the 70S ribosome.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4V4Z
 
 | | 70S Thermus thermophilous ribosome functional complex with mRNA and E- and P-site tRNAs at 4.5A. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.51 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
7QNN
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with inhibitor (surface entropy reduction mutant) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid C26-monooxygenase, ethyl 1-(cyclopentylmethyl)-5-pyridin-4-yl-indole-2-carboxylate | | Authors: | Snee, M, Tunnicliffe, R, Leys, D, Levy, C, Katariya, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
3KD3
 
 | | Crystal structure of a phosphoserine phosphohydrolase-like protein from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphoserine phosphohydrolase-like protein | | Authors: | Nocek, B, Zhou, M, Peterson, S, Anderson, W, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a phosphoserine phosphohydrolase-like protein from Francisella tularensis subsp. tularensis SCHUS4
TO BE PUBLISHED
|
|
4V67
 
 | | Crystal structure of a translation termination complex formed with release factor RF2. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Asahara, H, Lancaster, L, Laurberg, M, Hirschi, A, Noller, H.F. | | Deposit date: | 2008-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a translation termination complex formed with release factor RF2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3KDO
 
 | | Crystal structure of Type III Rubisco SP6 mutant complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
4V0S
 
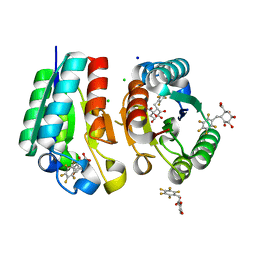 | | Crystal structure of Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (1R,2S,4S,5R)-2-(2,3,4,5,6-pentafluorophenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3,4-DIHYDROXY-2-[(2,3,4,5,6-PENTAFLUOROPHENYL)METHYL]BENZOIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Santiago, C, Lamb, H, Hawkins, A.R, Maneiro, M, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-09-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
3KEW
 
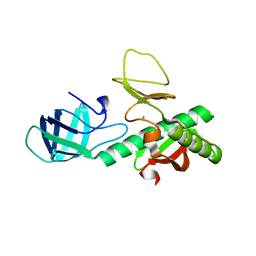 | | Crystal structure of probable alanyl-trna-synthase from Clostridium perfringens | | Descriptor: | DHHA1 domain protein, ZINC ION | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alanyl-trna-synthase from Clostridium perfringens
To be Published
|
|
4UQO
 
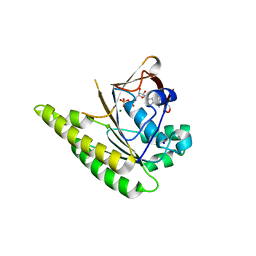 | | RADA C-TERMINAL ATPASE DOMAIN FROM PYROCOCCUS FURIOSUS BOUND TO ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA REPAIR AND RECOMBINATION PROTEIN RADA, MAGNESIUM ION, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2014-06-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
4USX
 
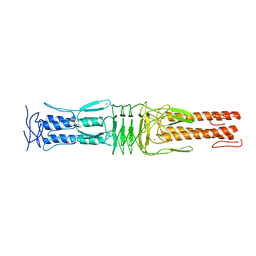 | | The Structure of the C-terminal YadA-like domain of BPSL2063 from Burkholderia pseudomallei | | Descriptor: | MAGNESIUM ION, TRIMERIC AUTOTRANSPORTER ADHESIN | | Authors: | Perletti, L, Gourlay, L.J, Peano, C, Pietrelli, A, DeBellis, G, Deantonio, C, Santoro, C, Sblattero, D, Bolognesi, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selecting Soluble/Foldable Protein Domains Through Single-Gene or Genomic Orf Filtering: Structure of the Head Domain of Burkholderia Pseudomallei Antigen Bpsl2063.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UYW
 
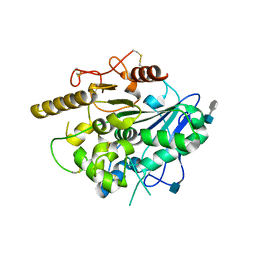 | |
4UZI
 
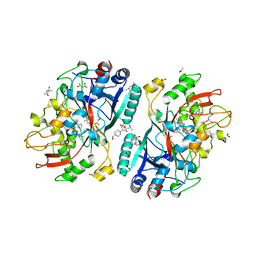 | | Crystal Structure of AauDyP Complexed with Imidazole | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Strittmatter, E, Liers, C, Ullrich, R, Hofrichter, M, Piontek, K, Plattner, D.A. | | Deposit date: | 2014-09-05 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Toolbox of Auricularia Auricula-Judae Dye-Decolorizing Peroxidase - Identification of Three New Potential Substrate-Interaction Sites.
Arch.Biochem.Biophys., 574, 2015
|
|
3KBX
 
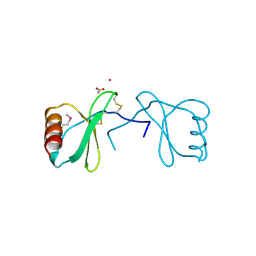 | |
4V6T
 
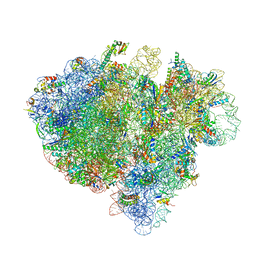 | | Structure of the bacterial ribosome complexed by tmRNA-SmpB and EF-G during translocation and MLD-loading | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Ramrath, D.J.F, Yamamoto, H, Rother, K, Wittek, D, Pech, M, Mielke, T, Loerke, J, Scheerer, P, Ivanov, P, Teraoka, Y, Shpanchenko, O, Nierhaus, K.H, Spahn, C.M.T. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | The complex of tmRNA-SmpB and EF-G on translocating ribosomes.
Nature, 485, 2012
|
|
4V4O
 
 | | Crystal Structure of the Chaperonin Complex Cpn60/Cpn10/(ADP)7 from Thermus Thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Shimamura, T, Koike-Takeshita, A, Yokoyama, K, Masui, R, Murai, N, Yoshida, M, Taguchi, H, Iwata, S. | | Deposit date: | 2004-05-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity
STRUCTURE, 12, 2004
|
|
5OPQ
 
 | | A 3,6-anhydro-D-galactosidase produced by Zobellia galactanivorans. This is an exo-lytic enzyme that hydrolyzes terminal 3,6-anhydro-D-galactose from the non-reducing end of carrageenan oligosaccharides. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6-anhydro-D-galactosidase, ... | | Authors: | Ficko-Blean, E, Michel, G, Czjzek, M. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria.
Nat Commun, 8, 2017
|
|
4V5V
 
 | | Structure of respiratory syncytial virus nucleocapsid protein, P1 crystal form | | Descriptor: | RESPIRATORY SYNCYTIAL VIRUS NUCLEOCAPSID PROTEIN, RNA | | Authors: | El Omari, K, Dhaliwal, B, Ren, J, Abrescia, N.G.A, Lockyer, M, Powell, K.L, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2011-05-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of Respiratory Syncytial Virus Nucleocapsid Protein from Two Crystal Forms: Details of Potential Packing Interactions in the Native Helical Form.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3KF6
 
 | | Crystal structure of S. pombe Stn1-ten1 complex | | Descriptor: | Protein stn1, Protein ten1 | | Authors: | Sun, J, Yu, E.Y, Yang, Y.T, Confer, L.A, Sun, S.H, Wan, K, Lue, N.F, Lei, M. | | Deposit date: | 2009-10-27 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres.
Genes Dev., 23, 2009
|
|
4V6U
 
 | | Promiscuous behavior of proteins in archaeal ribosomes revealed by cryo-EM: implications for evolution of eukaryotic ribosomes | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10P, ... | | Authors: | Armache, J.-P, Anger, A.M, Marquez, V, Frankenberg, S, Froehlich, T, Villa, E, Berninghausen, O, Thomm, M, Arnold, G.J, Beckmann, R, Wilson, D.N. | | Deposit date: | 2012-08-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Promiscuous behaviour of archaeal ribosomal proteins: Implications for eukaryotic ribosome evolution.
Nucleic Acids Res., 41, 2013
|
|
4V8D
 
 | | Structure analysis of ribosomal decoding (cognate tRNA-tyr complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
