6A33
 
 | | Binding and Enhanced Binding between Key Immunity Proteins TRAF6 and TIFA | | Descriptor: | 15-mer peptide from TRAF-interacting protein with FHA domain-containing protein A, TNF receptor-associated factor 6 | | Authors: | Huang, W.C, Liao, J.H, Hsiao, T.C, Maestre-Reyna, M, Bessho, Y, Tsai, M.D. | | Deposit date: | 2018-06-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding and Enhanced Binding between Key Immunity Proteins TRAF6 and TIFA.
Chembiochem, 20, 2019
|
|
6ABT
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
1AAY
 
 | | ZIF268 ZINC FINGER-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3'), PROTEIN (ZIF268 ZINC FINGER PEPTIDE), ... | | Authors: | Elrod-Erickson, M, Rould, M.A, Pabo, C.O. | | Deposit date: | 1997-01-18 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions.
Structure, 4, 1996
|
|
6R6N
 
 | |
8TNR
 
 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 2 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
6R9T
 
 | | Cryo-EM structure of autoinhibited human talin-1 | | Descriptor: | Talin-1 | | Authors: | Dedden, D, Schumacher, S, Zacharias, M, Biertumpfel, C, Mizuno, N. | | Deposit date: | 2019-04-04 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | The Architecture of Talin1 Reveals an Autoinhibition Mechanism.
Cell, 179, 2019
|
|
5ZRQ
 
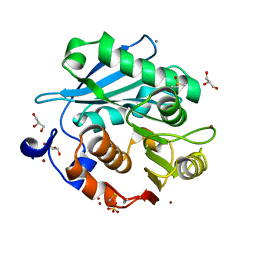 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in Zn(2+)-bound state | | Descriptor: | Alpha/beta hydrolase family protein, CALCIUM ION, GLYCEROL, ... | | Authors: | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
6RBS
 
 | |
6RBZ
 
 | |
6RBP
 
 | |
6AAD
 
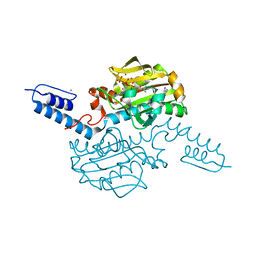 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with mTmdZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N6-[({3-[3-(trifluoromethyl)-3H-diaziren-3-yl]phenyl}methoxy)carbonyl]-L-lysine, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
8R2I
 
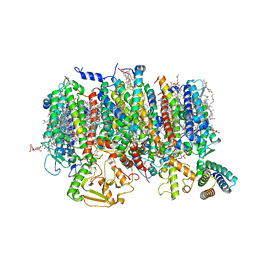 | | Cryo-EM Structure of native Photosystem II assembly intermediate from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Fadeeva, M, Klaiman, D, Kandiah, E, Nelson, N. | | Deposit date: | 2023-11-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of native photosystem II assembly intermediate from Chlamydomonas reinhardtii .
Front Plant Sci, 14, 2023
|
|
6RBY
 
 | |
5ZT4
 
 | |
6ACG
 
 | |
5ZVX
 
 | |
6R2L
 
 | | NYBR1-A2-SLSKILDTV | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sami, M. | | Deposit date: | 2019-03-18 | | Release date: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NYBR1-A2-SLSKILDTV
To Be Published
|
|
6R2D
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD after soaking with succinylphosphonate phosphonoethyl ester, followed by temperature increase | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-[ethoxy(oxidanyl)phosphoryl]-4-oxidanyl-butanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6R2C
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD after soaking with succinylphosphonate phosphonoethyl ester (PESP) | | Descriptor: | 4-[ethoxy(oxidanyl)phosphoryl]-4-oxidanylidene-butanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6R2B
 
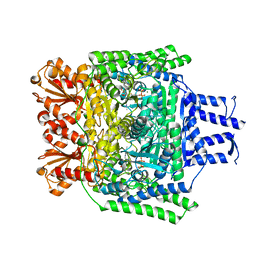 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD after soaking with succinylphosphonate | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-oxidanyl-4-phosphono-butanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
6A0J
 
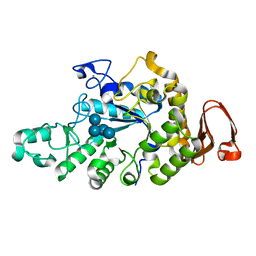 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, complex with Cyclic alpha-maltosyl-(1-->6)-maltose | | Descriptor: | CALCIUM ION, Cyclic alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cyclic maltosyl-maltose hydrolase | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
6R6M
 
 | |
5ZZN
 
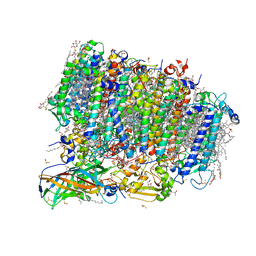 | | Crystal structure of photosystem II from an SQDG-deficient mutant of Thermosynechococcus elongatus | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Nakajima, Y, Umena, Y, Nagao, R, Endo, K, Kobayashi, K, Akita, F, Suga, M, Wada, H, Noguchi, T, Shen, J.R. | | Deposit date: | 2018-06-03 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thylakoid membrane lipid sulfoquinovosyl-diacylglycerol (SQDG) is required for full functioning of photosystem II inThermosynechococcus elongatus.
J. Biol. Chem., 293, 2018
|
|
6A2U
 
 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Glucose dehydrogenase, ... | | Authors: | Yoshida, H, Kojima, K, Yoshimatsu, K, Shiota, M, Yamazaki, T, Ferri, S, Tsugawa, W, Kamitori, S, Sode, K. | | Deposit date: | 2018-06-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the direct electron transfer-type FAD glucose dehydrogenase catalytic subunit complexed with a hitchhiker protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6ABI
 
 | | The apo-structure of D-lactate dehydrogenase from Fusobacterium nucleatum | | Descriptor: | D-lactate dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
