8H6Q
 
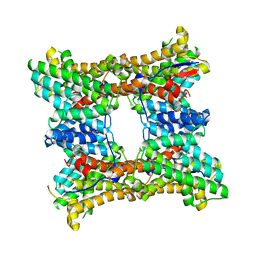 | | Class I sesquiterpene synthase BCBOT2 (apo) | | Descriptor: | Presilphiperfolan-8-beta-ol synthase | | Authors: | Lou, T.T, Ma, M. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Three Sesquiterpene Synthases for the Biosynthesis of Tricyclic Sesquiterpenes and Chemical Space Expansion by Structure-Based Mutagenesis.
J.Am.Chem.Soc., 2023
|
|
8H6U
 
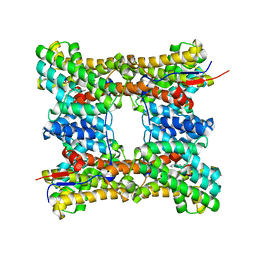 | | Class I sesquiterpene synthase BCBOT2 (complex) | | Descriptor: | MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, PYROPHOSPHATE 2-, ... | | Authors: | Lou, T.T, Ma, M. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural Insights into Three Sesquiterpene Synthases for the Biosynthesis of Tricyclic Sesquiterpenes and Chemical Space Expansion by Structure-Based Mutagenesis.
J.Am.Chem.Soc., 2023
|
|
8H4P
 
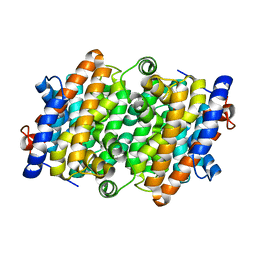 | | class I sesquiterpene synthase | | Descriptor: | Longiborneol synthase CLM1, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Li, A.N, Lou, T.T, Ma, M. | | Deposit date: | 2022-10-11 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into Three Sesquiterpene Synthases for the Biosynthesis of Tricyclic Sesquiterpenes and Chemical Space Expansion by Structure-Based Mutagenesis.
J.Am.Chem.Soc., 2023
|
|
8H72
 
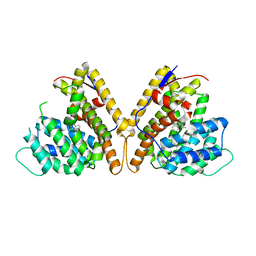 | | Class I sesquiterpene synthase DbPROS (complex) | | Descriptor: | Delta(6)-protoilludene synthase K435DRAFT_659367, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium | | Authors: | Lou, T.T, Ma, M. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Insights into Three Sesquiterpene Synthases for the Biosynthesis of Tricyclic Sesquiterpenes and Chemical Space Expansion by Structure-Based Mutagenesis.
J.Am.Chem.Soc., 2023
|
|
5YP2
 
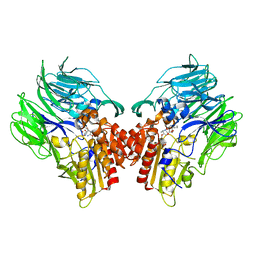 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 | | Descriptor: | (2S,5R)-1-[2-[[1-(hydroxymethyl)cyclopentyl]amino]ethanoyl]pyrrolidine-2,5-dicarbonitrile, Dipeptidyl aminopeptidase 4, GLYCEROL | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
8H25
 
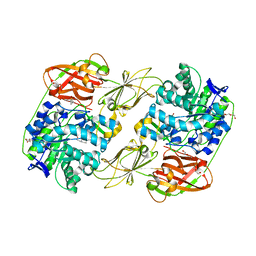 | | Lacticaseibacillus casei GH35 beta-galactosidase LBCZ_0230 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saburi, W, Ota, T, Kato, K, Tagami, T, Yamashita, K, Yao, M, Mori, H. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Function and Structure of Lacticaseibacillus casei GH35 beta-Galactosidase LBCZ_0230 with High Hydrolytic Activity to Lacto- N -biose I and Galacto- N -biose.
J Appl Glycosci (1999), 70, 2023
|
|
7CW2
 
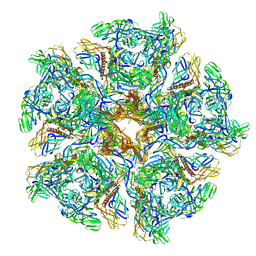 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-263 (subregion around icosahedral 5-fold vertex) | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8VSK
 
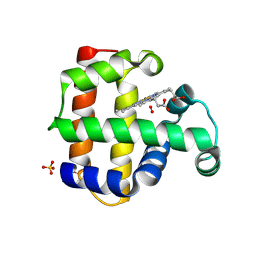 | | Crystal structure of Dehaloperoxidase A in complex with substrate 2,4-dibromophenol | | Descriptor: | 2,4-bis(bromanyl)phenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VKD
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VZR
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-bromo-o-cresol | | Descriptor: | 4-bromo-2-methylphenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
7CVZ
 
 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-263 | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8VKC
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrophenol | | Descriptor: | Dehaloperoxidase A, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
7CVY
 
 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-124 | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8VWR
 
 | | Structure of steroid hydratase from Comamonas testosteroni | | Descriptor: | Acyl dehydratase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schroeter, K.L, Forrester, T.J.B, Kimber, M.S, Seah, S.Y.K. | | Deposit date: | 2024-02-02 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Shy is a proteobacterial steroid hydratase which catalyzes steroid side chain degradation without requiring a catalytically inert partner domain.
J.Biol.Chem., 300, 2024
|
|
8GVD
 
 | | SARS-CoV-2 Mpro in complex with D-4-38 | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, 3C-like proteinase nsp5 | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 Mpro in complex with D-4-38
To Be Published
|
|
8GX3
 
 | | The crystal structure of human Calpain-1 protease core in complex with 14c | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of human Calpain-1 protease core in complex with 14c
To Be Published
|
|
5YJI
 
 | | Co-crystal structure of Mouse Nicotinamide N-methyltransferase (NNMT) with small molecule analog of Nicotinamide | | Descriptor: | 6-methoxy-1-methyl-2H-pyridine-3-carboxamide, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Birudukota, S, Swaminathan, S, Thakur, M.K, Parveen, R, Kandan, S, Hallur, M.S, Rajagopal, S, Ruf, S, Dhakshinamoorthy, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 8, 2018
|
|
8GVY
 
 | | SARS CoV-2 Mpro in complex with D-3-149 | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, 3C-like proteinase nsp5 | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SARS CoV-2 Mpro in complex with D-3-149
To Be Published
|
|
8GX2
 
 | | The crystal structure of human CtsL in complex with 14c | | Descriptor: | DIMETHYL SULFOXIDE, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human CtsL in complex with 14a
To Be Published
|
|
7WCQ
 
 | | Crystal structure of HIV-1 protease in complex with lactam derivative 1 | | Descriptor: | (3R,4R)-3-[(4-fluorophenyl)methyl]-1-[(4-methoxyphenyl)methyl]-3-(4-methylsulfonylphenyl)-4-oxidanyl-pyrrolidin-2-one, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakasa-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
7WBS
 
 | | Crystal structure of HIV-1 protease in complex with lactam derivative 2 | | Descriptor: | (3~{R},4~{R})-1-[(4-methoxyphenyl)methyl]-3-(3-methylbutyl)-3-[4-methylsulfonyl-2-[(2~{S})-1-oxidanylpropan-2-yl]oxy-phenyl]-4-oxidanyl-pyrrolidin-2-one, GLYCEROL, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakabayashi-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
8GXI
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14c | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The crystal structure of SARS-CoV-2 main protease in complex with 14c
To Be Published
|
|
5YJF
 
 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with small molecule analog of Nicotinamide | | Descriptor: | 6-methoxy-1-methyl-2H-pyridine-3-carboxamide, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Birudukota, S, Thakur, M.K, Parveen, R, Kandan, S, Hallur, M.S, Rajagopal, S, Ruf, S, Dhakshinamoorthy, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 8, 2018
|
|
8GWJ
 
 | |
8GYE
 
 | |
