8SHQ
 
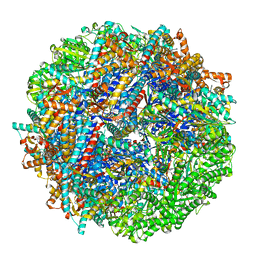 | | CCT G beta 5 complex closed state 12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHF
 
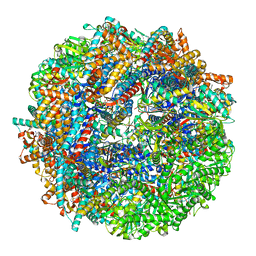 | | CCT-G beta 5 complex closed state 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
7OCI
 
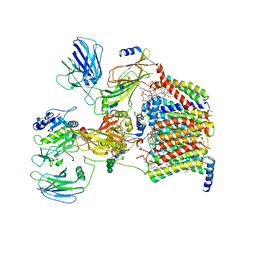 | | Cryo-EM structure of yeast Ost6p containing oligosaccharyltransferase complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wild, R, Neuhaus, J.D, Eyring, J, Irobalieva, R.N, Kowal, J, Lin, C.W, Locher, K.P, Aebi, M. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Functional analysis of Ost3p and Ost6p containing yeast oligosaccharyltransferases.
Glycobiology, 31, 2021
|
|
7NRG
 
 | |
4P1X
 
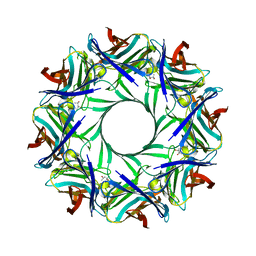 | | Crystal structure of staphylococcal LUK prepore | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Gamma-hemolysin component B, Gamma-hemolysin component C | | Authors: | Yamashita, D, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2014-02-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of transmembrane beta-barrel formation of staphylococcal pore-forming toxins.
Nat Commun, 5, 2014
|
|
7NS7
 
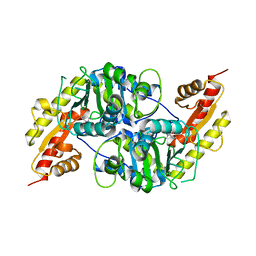 | |
3B3K
 
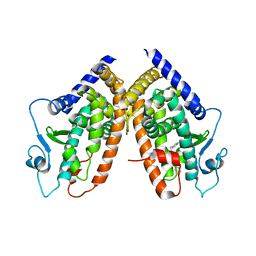 | | Crystal structure of the complex between PPARgamma and the full agonist LT175 | | Descriptor: | (2S)-2-(biphenyl-4-yloxy)-3-phenylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Mazza, F, Loiodice, F, Fracchiolla, G, Crestani, M, Godio, C. | | Deposit date: | 2007-10-22 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Peroxisome Proliferator-Activated Receptor gamma (PPARgamma) Ligand Binding Domain Complexed with a Novel Partial Agonist: A New Region of the Hydrophobic Pocket Could Be Exploited for Drug Design
J.Med.Chem., 51, 2008
|
|
7O05
 
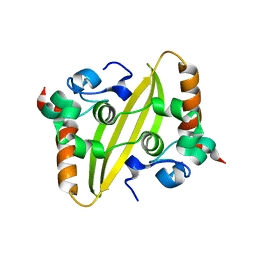 | |
6YE4
 
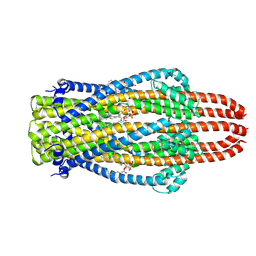 | | Structure of ExbB pentamer from Serratia marcescens by single particle cryo electron microscopy | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Biopolymer transport protein ExbB | | Authors: | Biou, V, Delepelaire, P, Coureux, P.D, Chami, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and molecular determinants for the interaction of ExbB from Serratia marcescens and HasB, a TonB paralog.
Commun Biol, 5, 2022
|
|
7O04
 
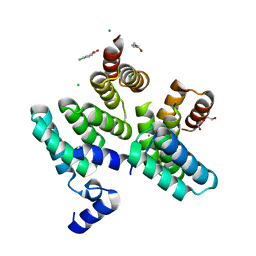 | |
7O36
 
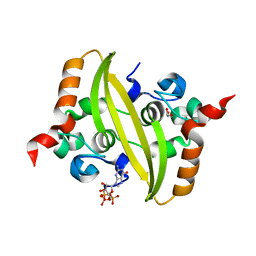 | |
6Y7A
 
 | | X-ray structure of the Haloalkane dehalogenase HaloTag7 labeled with a chloroalkane-tetramethylrhodamine fluorophore substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-02-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
5YP6
 
 | | RORgamma (263-509) complexed with SRC2 and Compound 6 | | Descriptor: | N-[3'-cyano-4'-(2-methylpropyl)-2-(trifluoromethyl)biphenyl-4-yl]-2-[4-(ethylsulfonyl)phenyl]acetamide, Nuclear receptor ROR-gamma, SRC2 | | Authors: | Gao, M, Cai, W. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | From ROR gamma t Agonist to Two Types of ROR gamma t Inverse Agonists
ACS Med Chem Lett, 9, 2018
|
|
5Y0B
 
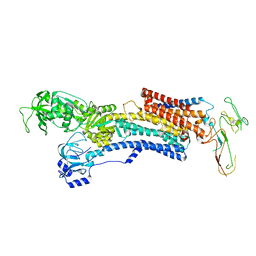 | | PIG GASTRIC H+,K+ - ATPASE IN COMPLEX with BYK99 | | Descriptor: | Potassium-transporting ATPase alpha chain 1, Potassium-transporting ATPase subunit beta | | Authors: | Abe, K, Shimokawa, J, Natio, M, Munson, K, Vagin, O, Sachs, G, Suzuki, H, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2017-07-16 | | Release date: | 2017-08-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (6.7 Å) | | Cite: | The cryo-EM structure of gastric H(+),K(+)-ATPase with bound BYK99, a high-affinity member of K(+)-competitive, imidazo[1,2-a]pyridine inhibitors
Sci Rep, 7, 2017
|
|
6Y7B
 
 | | X-ray structure of the Haloalkane dehalogenase HaloTag7 labeled with a chloroalkane-carbopyronine fluorophore substrate | | Descriptor: | 4-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]-2-[3-(dimethylamino)-6-(dimethyl-$l^{4}-azanylidene)-10,10-dimethyl-anthracen-9-yl]benzoic acid, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Tarnawski, M, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-02-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
7O1N
 
 | |
3C8G
 
 | | Crystal structure of a possible transciptional regulator YggD from Shigella flexneri 2a str. 2457T | | Descriptor: | ACETATE ION, Putative transcriptional regulator | | Authors: | Tan, K, Borovilos, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-12 | | Release date: | 2008-02-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
7O3D
 
 | | Cooperation between the intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Kasiliauskaite, A, Kubicek, K, Klumpler, T, Zanova, M, Zapletal, D, Novacek, J, Stefl, R. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res., 50, 2022
|
|
7O35
 
 | |
4PBE
 
 | | Phosphotriesterase Variant Rev6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-revR6, ... | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-04-12 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat. Chem. Biol., 12, 2016
|
|
7O6B
 
 | | Cooperation between the intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Kasiliauskaite, A, Kubicek, K, Klumpler, T, Zanova, M, Zapletal, D, Novacek, J, Stefl, R. | | Deposit date: | 2021-04-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res., 50, 2022
|
|
4P1Y
 
 | | Crystal structure of staphylococcal gamma-hemolysin prepore | | Descriptor: | Gamma-hemolysin component A, Gamma-hemolysin component B | | Authors: | Yamashita, D, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2014-02-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | Molecular basis of transmembrane beta-barrel formation of staphylococcal pore-forming toxins.
Nat Commun, 5, 2014
|
|
8SKS
 
 | | human liver mitochondrial Superoxide dismutase [Mn] | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Zhang, Z, Tringides, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
8SKR
 
 | | human liver mitochondrial Aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, mitochondrial, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhang, Z, Tringides, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
8S2X
 
 | |
