3G3T
 
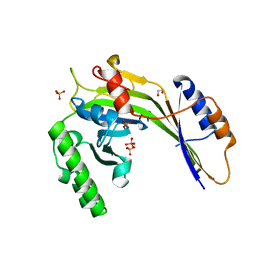 | | Crystal structure of a eukaryotic polyphosphate polymerase in complex with orthophosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Vacuolar transporter chaperone 4 | | Authors: | Lenherr, E.D, Hothorn, M, Scheffzek, K. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase.
Science, 324, 2009
|
|
4A38
 
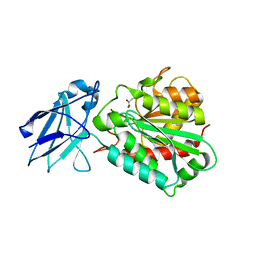 | | METALLO-CARBOXYPEPTIDASE FROM PSEUDOMONAS AUREGINOSA IN COMPLEX WITH L-BENZYLSUCCINIC ACID | | Descriptor: | L-BENZYLSUCCINIC ACID, METALLO-CARBOXYPEPTIDASE, ZINC ION | | Authors: | Otero, A, Rodriguez de la Vega, M, Tanco, S.M, Lorenzo, J, Aviles, F.X, Reverter, D. | | Deposit date: | 2011-09-30 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Novel Structure of a Cytosolic M14 Metallocarboxypeptidase (Ccp) from Pseudomonas Aeruginosa: A Model for Mammalian Ccps.
Faseb J., 26, 2012
|
|
4AL1
 
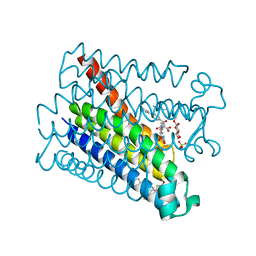 | | Crystal structure of Human PS-1 GSH-analog complex | | Descriptor: | L-gamma-glutamyl-S-(2-biphenyl-4-yl-2-oxoethyl)-L-cysteinylglycine, PALMITIC ACID, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Sjogren, T, Nord, J, Ek, M, Johansson, P, Liu, G, Geschwindner, S. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Microsomal Prostaglandin E2 Synthase Provides Insight Into Diversity in the Mapeg Superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4A60
 
 | | Crystal structure of human testis-specific fatty acid binding protein 9 (FABP9) | | Descriptor: | 1,2-ETHANEDIOL, FATTY ACID-BINDING PROTEIN 9 TESTIS LIPID-BINDING PROTEIN, TLBP, ... | | Authors: | Muniz, J.R.C, Kiyani, W, Shrestha, L, Froese, D.S, Krojer, T, Vollmar, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, von Delft, F, Yue, W.W. | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Crystal Structure of the Human Fabp9A
To be Published
|
|
2ODI
 
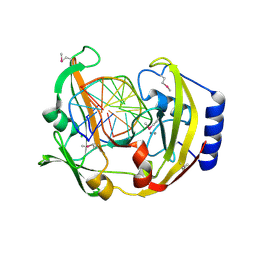 | | Restriction Endonuclease BCNI-Cognate DNA Substrate Complex | | Descriptor: | 5'-D(*AP*AP*CP*CP*CP*GP*GP*AP*GP*AP*C)-3', 5'-D(*CP*TP*CP*CP*GP*GP*GP*TP*TP*GP*T)-3', CALCIUM ION, ... | | Authors: | Sokolowska, M, Kaus-Drobek, M, Czapinska, H, Tamulaitis, G, Szczepanowski, R.H, Urbanke, C, Siksnys, V, Bochtler, M. | | Deposit date: | 2006-12-22 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Monomeric restriction endonuclease BcnI in the apo form and in an asymmetric complex with target DNA.
J.Mol.Biol., 369, 2007
|
|
478D
 
 | |
4AM9
 
 | | CRYSTAL STRUCTURE OF THE YERSINIA ENTEROCOLITICA TYPE III SECRETION CHAPERONE SYCD IN COMPLEX WITH A PEPTIDE OF THE TRANSLOCATOR YOPD | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHAPERONE SYCD, SULFATE ION, ... | | Authors: | Schreiner, M, Niemann, H.H. | | Deposit date: | 2012-03-08 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Yersinia Enterocolitica Type III Secretion Chaperone Sycd in Complex with a Peptide of the Minor Translocator Yopd
Bmc Struct.Biol., 12, 2012
|
|
3KAT
 
 | | Crystal Structure of the CARD domain of the human NLRP1 protein, Northeast Structural Genomics Consortium Target HR3486E | | Descriptor: | NACHT, LRR and PYD domains-containing protein 1 | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR3486E
To be Published
|
|
6F2X
 
 | | Structural characterization of the Mycobacterium tuberculosis Protein Tyrosine Kinase A (PtkA) | | Descriptor: | Protein Tyrosine Kinase A | | Authors: | Niesteruk, A, Jonker, H.R.A, Sreeramulu, S, Richter, C, Hutchison, M, Linhard, V, Schwalbe, H. | | Deposit date: | 2017-11-27 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The domain architecture of PtkA, the first tyrosine kinase fromMycobacterium tuberculosis, differs from the conventional kinase architecture.
J. Biol. Chem., 293, 2018
|
|
4AAS
 
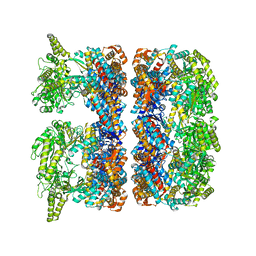 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
4A5O
 
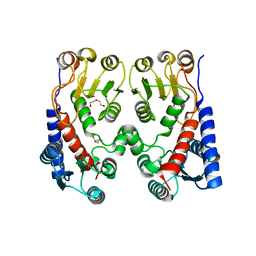 | | Crystal structure of Pseudomonas aeruginosa N5, N10- methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) | | Descriptor: | BIFUNCTIONAL PROTEIN FOLD, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Eadsforth, T.C, Gardiner, M, Maluf, F.V, McElroy, S, James, D, Frearson, J, Gray, D, Hunter, W.N. | | Deposit date: | 2011-10-26 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Assessment of Pseudomonas Aeruginosa N(5),N(10)-Methylenetetrahydrofolate Dehydrogenase - Cyclohydrolase as a Potential Antibacterial Drug Target.
Plos One, 7, 2012
|
|
4AKJ
 
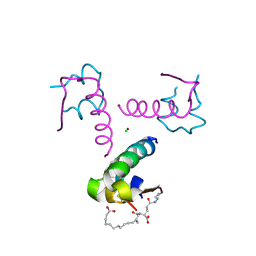 | | Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Steensgaard, D.B, Schluckebier, G, Strauss, H.M, Norrman, M, Thomsen, J.K, Friderichsen, A.V, Havelund, S, Jonassen, I. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Ligand Controlled Assembly of Hexamers, Dihexamers, and Linear Multihexamer Structures by the Engineered Acylated Insulin Degludec.
Biochemistry, 52, 2013
|
|
4BJP
 
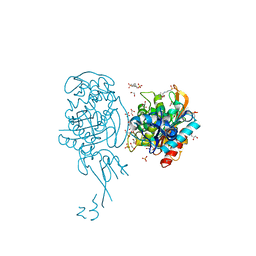 | | Crystal structure of E. coli penicillin binding protein 3 | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, ... | | Authors: | Sauvage, E, Joris, M, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2013-04-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 3 (Pbp3) from Escherichia Coli.
Plos One, 9, 2014
|
|
1GTV
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE-5'-DIPHOSPHATE (TDP) | | Descriptor: | ACETATE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ursby, T, Weik, M, Fioravanti, E, Delarue, M, Goeldner, M, Bourgeois, D. | | Deposit date: | 2002-01-21 | | Release date: | 2002-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cryophotolysis of Caged Compounds: A Technique for Trapping Intermediate States in Protein Crystals
Acta Crystallogr.,Sect.D, 58, 2002
|
|
460D
 
 | |
1QGZ
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 78 REPLACED BY ASP (L78D) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
4AE9
 
 | | Structure and function of the Human Sperm-Specific Isoform of Protein Kinase A (PKA) Catalytic Subunit C alpha 2 | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA | | Authors: | Hereng, T.H, Backe, P.H, Kahmann, J, Scheich, C, Bjoras, M, Skalhegg, B.S, Rosendal, K.R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of the Human Sperm-Specific Isoform of Protein Kinase a (Pka) Catalytic Subunit Calpha2
J.Struct.Biol., 178, 2012
|
|
1H85
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH VAL 136 REPLACED BY LEU (V136L) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a Cluster of Hydrophobic Residues Near the Fad Cofactor in Anabaena Pcc 7119 Ferredoxin-Nadp+ Reductase for Optimal Complex Formation and Electron Transfer to Ferredoxin
J.Biol.Chem., 276, 2001
|
|
4A2O
 
 | | STRUCTURE OF THE HUMAN EOSINOPHIL CATIONIC PROTEIN IN COMPLEX WITH SULFATE ANIONS | | Descriptor: | EOSINOPHIL CATIONIC PROTEIN, SULFATE ION | | Authors: | Boix, E, Pulido, D, Moussaoui, M, Nogues, V, Russi, S. | | Deposit date: | 2011-09-28 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The Sulfate-Binding Site Structure of the Human Eosinophil Cationic Protein as Revealed by a New Crystal Form.
J.Struct.Biol., 179, 2012
|
|
3X40
 
 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium chloride | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
4X1S
 
 | | The crystal structure of mupain-1-16-D9A in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1-16, piperidine-1-carboximidamide | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-10-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
3FRI
 
 | | Structure of the 16S rRNA methylase RmtB, I222 | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Dupechez, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-08 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
3ZGP
 
 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase acylated by ertapenem | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Triboulet, S, Dubee, V, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-18 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
3ZS5
 
 | | Structural basis for kinase selectivity of three clinical p38alpha inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MITOGEN-ACTIVATED PROTEIN KINASE 14, ... | | Authors: | Azevedo, R, van Zeeland, M, Raaijmakers, H.C.A, Kazemier, B, Oubrie, A. | | Deposit date: | 2011-06-23 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure of p38 alpha bound to TAK-715: comparison with three classic inhibitors.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
3ZM5
 
 | | CRYSTAL STRUCTURE OF MURF LIGASE IN COMPLEX WITH CYANOTHIOPHENE INHIBITOR | | Descriptor: | 2,4-bis(chloranyl)-N-[3-cyano-6-[(4-hydroxyphenyl)methyl]-5,7-dihydro-4H-thieno[2,3-c]pyridin-2-yl]-5-morpholin-4-ylsulfonyl-benzamide, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | Authors: | Hrast, M, Turk, S, Sosic, I, Knez, D, Randall, C.P, Barreteau, H, Contreras-Martel, C, Dessen, A, ONeill, A.J, Mengin-Lecreulx, D, Blanot, D, Gobec, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure-Activity Relationships of New Cyanothiophene Inhibitors of the Essential Peptidoglycan Biosynthesis Enzyme Murf.
Eur.J.Med.Chem., 66C, 2013
|
|
