1WX7
 
 | | Solution Structure of the N-terminal Ubiquitin-like Domain in the Human Ubiquilin 3 (UBQLN3) | | Descriptor: | Ubiquilin 3 | | Authors: | Zhao, C, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the N-terminal Ubiquitin-like Domain in the Human Ubiquilin 3 (UBQLN3)
To be Published
|
|
1WY0
 
 | |
1WYP
 
 | | Solution structure of the CH domain of human Calponin 1 | | Descriptor: | Calponin 1 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-15 | | Release date: | 2005-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain of human Calponin 1
To be Published
|
|
7ZC8
 
 | |
4G1R
 
 | | Crystal structure of anti-HIV actinohivin in complex with alphs-1,2-mannobiose (Form II) | | Descriptor: | Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-07-11 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Matured structure of anti-HIV lectin actinohivin in complex with alpha-1,2-mannobiose
To be Published
|
|
4FUL
 
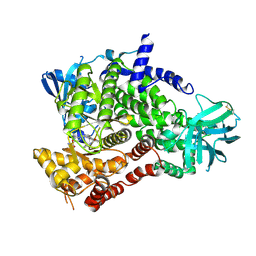 | | PI3 Kinase Gamma bound to a pyrmidine inhibitor | | Descriptor: | 4-({4-[3-(piperidin-1-ylcarbonyl)phenyl]pyrimidin-2-yl}amino)benzenesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Gopalsamy, A, Bennett, E.M, Shi, M, Zhang, W.G, Bard, J, Yu, K. | | Deposit date: | 2012-06-28 | | Release date: | 2012-10-17 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification of pyrimidine derivatives as hSMG-1 inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1X0J
 
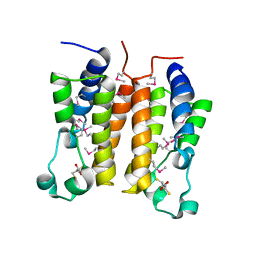 | | Crystal structure analysis of the N-terminal bromodomain of human Brd2 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-03-23 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for acetylated histone H4 recognition by the human Brd2 bromodomain
To be Published
|
|
4G23
 
 | |
2RLI
 
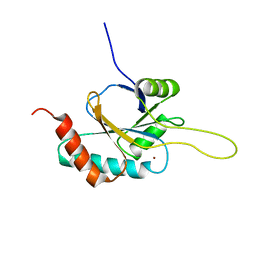 | | Solution structure of Cu(I) human Sco2 | | Descriptor: | COPPER (I) ION, SCO2 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-baffoni, S, Gerothanassis, I.P, Leontari, I, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural Characterization of Human SCO2
Structure, 15, 2007
|
|
4G3V
 
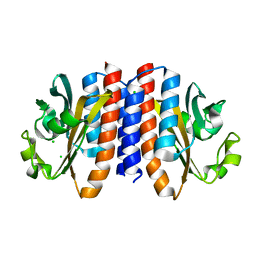 | | Crystal structure of A. Aeolicus nlh2 gaf domain in an inactive state | | Descriptor: | CHLORIDE ION, Transcriptional regulator nlh2 | | Authors: | Batchelor, J.D, Lee, P, Wang, A, Doucleff, M, Wemmer, D.E. | | Deposit date: | 2012-07-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism of GAF-regulated delta(54) activators from Aquifex aeolicus
J.Mol.Biol., 425, 2013
|
|
2REX
 
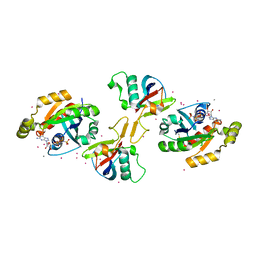 | | Crystal structure of the effector domain of PLXNB1 bound with Rnd1 GTPase | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Tong, Y, Tempel, W, Shen, L, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-27 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the effector domain of PLXNB1 bound with Rnd1 GTPase.
To be Published
|
|
1WGS
 
 | | Solution Structure of the Tudor Domain from Mouse Hypothetical Protein Homologous to Histone Acetyltransferase | | Descriptor: | MYST histone acetyltransferase 1 | | Authors: | Li, H, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Tudor Domain from Mouse Hypothetical Protein Homologous to Histone Acetyltransferase
To be Published
|
|
1WHD
 
 | | Solution structure of the PDZ domain of RGS3 | | Descriptor: | regulator of G-protein signaling 3 | | Authors: | Nakanishi, T, Nemoto, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of RGS3
To be Published
|
|
2ROZ
 
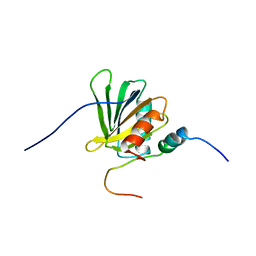 | | Structure of the C-terminal PID Domain of Fe65L1 Complexed with the Cytoplasmic Tail of APP Reveals a Novel Peptide Binding Mode | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 2, peptide from Amyloid beta A4 protein | | Authors: | Li, H, Koshiba, S, Tochio, N, Watanabe, S, Harada, T, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
4ZDO
 
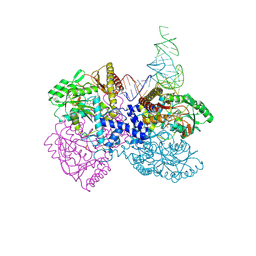 | |
1WJI
 
 | | Solution Structure of the UBA Domain of Human Tudor Domain Containing Protein 3 | | Descriptor: | Tudor domain containing protein 3 | | Authors: | Kamatari, Y.O, Tochio, N, Nakanishi, T, Miyamoto, K, Li, H, Kobayashi, N, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the UBA Domain of Human Tudor Domain Containing Protein 3
To be Published
|
|
4ZEG
 
 | | Crystal structure of TTK kinase domain in complex with a pyrazolopyrimidine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dual specificity protein kinase TTK, ... | | Authors: | Qiu, W, Plotnikova, O, Feher, M, Awrey, D.E, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2015-04-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of TTK kinase domain in complex with a pyrazolopyrimidine inhibitor.
To Be Published
|
|
1WJS
 
 | | Solution structure of the first mbt domain from human KIAA1798 protein | | Descriptor: | KIAA1798 protein | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first mbt domain from human KIAA1798 protein
To be Published
|
|
3MGF
 
 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 7.5 | | Descriptor: | Fluorescent protein | | Authors: | Ebisawa, T, Yamamura, A, Ohtsuka, J, Kameda, Y, Hayakawa, K, Nagata, K, Tanokura, M. | | Deposit date: | 2010-04-06 | | Release date: | 2011-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 7.5
To be Published
|
|
1WW9
 
 | | Crystal structure of the terminal oxygenase component of carbazole 1,9a-dioxygenase, a non-heme iron oxygenase system catalyzing the novel angular dioxygenation for carbazole and dioxin | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, terminal oxygenase component of carbazole | | Authors: | Nojiri, H, Ashikawa, Y, Noguchi, H, Nam, J.-W, Urata, M, Fujimoto, Z, Mizuno, H, Yoshida, T, Habe, H, Omori, T. | | Deposit date: | 2005-01-05 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the terminal oxygenase component of angular dioxygenase, carbazole 1,9a-dioxygenase
J.Mol.Biol., 351, 2005
|
|
1WLZ
 
 | | Crystal structure of DJBP fragment which was obtained by limited proteolysis | | Descriptor: | CAP-binding protein complex interacting protein 1 isoform a | | Authors: | Honbou, K, Suzuki, N, Horiuchi, M, Taira, T, Niki, T, Ariga, H, Inagaki, F. | | Deposit date: | 2004-07-01 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of DJBP Fragment which was obtained by Limited Proteolysis
To be Published
|
|
1WWM
 
 | | Crystal Structure of Conserved Hypothetical Protein TT2028 from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TT2028 | | Authors: | Mizohata, E, Ushikoshi-Nakayama, R, Terada, T, Murayama, K, Sakai, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-07 | | Release date: | 2005-07-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of TT2028 from an Extremely Thermophilic Bacterium Thermus thermophilus HB8
To be Published
|
|
2RIO
 
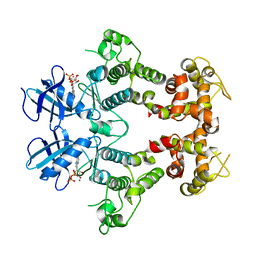 | | Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation of non-conventional splicing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, STRONTIUM ION, ... | | Authors: | Lee, K.P, Dey, M, Neculai, D, Cao, C, Dever, T.E, Sicheri, F. | | Deposit date: | 2007-10-12 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the dual enzyme ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing.
Cell(Cambridge,Mass.), 132, 2008
|
|
1WWV
 
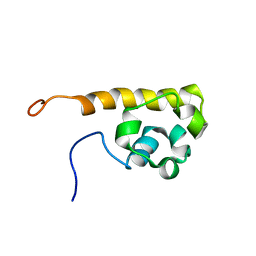 | | Solution structure of the SAM domain of human connector enhancer of KSR-like protein CNK1 | | Descriptor: | Connector enhancer of kinase suppressor of ras 1 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM domain of human connector enhancer of KSR-like protein CNK1
To be Published
|
|
4GAP
 
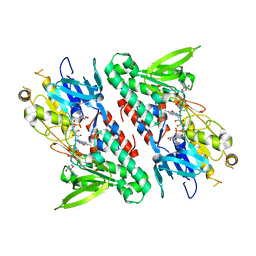 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with NAD+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
