5T4K
 
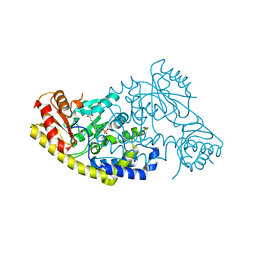 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | (4S)-5-fluoro-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]pentanoic acid, HTH-type transcriptional regulatory protein GabR | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8EAF
 
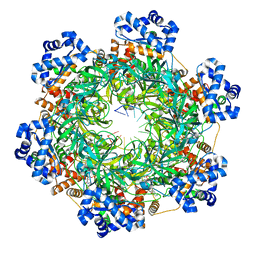 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 12-mer oligo-dT. Class 1 | | Descriptor: | 12-mer oligo dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
5T4M
 
 | |
1KWS
 
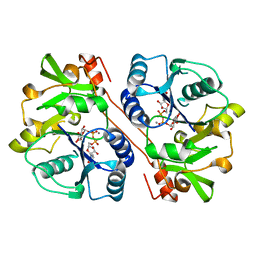 | | CRYSTAL STRUCTURE OF BETA1,3-GLUCURONYLTRANSFERASE I IN COMPLEX WITH THE ACTIVE UDP-GLCUA DONOR | | Descriptor: | BETA-1,3-GLUCURONYLTRANSFERASE 3, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Pedersen, L.C, Darden, T.A, Negishi, M. | | Deposit date: | 2002-01-30 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta 1,3-glucuronyltransferase I in complex with active donor substrate UDP-GlcUA.
J.Biol.Chem., 277, 2002
|
|
5T4O
 
 | | Autoinhibited E. coli ATP synthase state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Smits, C, Wong, A.S.W, Ishmukhametov, R, Stock, D, Sandin, S, Stewart, A.G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-EM structures of the autoinhibitedE. coliATP synthase in three rotational states.
Elife, 5, 2016
|
|
5KED
 
 | | Structure of the 2.65 Angstrom P2(1) crystal of K. pneumonia MrkH | | Descriptor: | Flagellar brake protein YcgR | | Authors: | Schumacher, M. | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | To be released:
Structures of K. pneumonia MrkH: dual utilization of the PilZ fold for c-di-GMP and DNA binding by a novel activator of biofilm genes
To Be Published
|
|
5G4K
 
 | | Phloroglucinol reductase from Clostridium sp. apo-form | | Descriptor: | OXIDOREDUCTASE, SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY PROTEIN | | Authors: | Conradt, D, Hermann, B, Gerhardt, S, Einsle, O, Mueller, M. | | Deposit date: | 2016-05-13 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biocatalytic Properties and Structural Analysis of Phloroglucinol Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5T6P
 
 | |
5G50
 
 | |
5T74
 
 | | Human carboanhydrase F131C_C206S double mutant in complex with 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[2,5-bis(oxidanylidene)pyrrol-1-yl]-~{N}-(4-sulfamoylphenyl)ethanamide, 4-(HYDROXYMERCURY)BENZOIC ACID, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
1BTT
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
5T6I
 
 | | CRYSTAL STRUCTURE OF TGCDPK1 FROM TOXOPLASMA GONDII COMPLEXED WITH 5GB | | Descriptor: | 4-(3-methylphenyl)-5-(1,5-naphthyridin-2-yl)-1,3-thiazol-2-amine, Calmodulin-domain protein kinase 1 | | Authors: | Jiang, D.Q, Tempel, W, Walker, J.R, El Bakkouri, M, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lovato, D.V, Osman, K.T, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of CDPK1 from toxoplasma gondii complexed with SGC-1-19
to be published
|
|
2O8P
 
 | | Crystal structure of a putative 14-3-3 protein from Cryptosporidium parvum, cgd7_2470 | | Descriptor: | 14-3-3 domain containing protein | | Authors: | Dong, A, Lew, J, Wasney, G, Lin, L, Hassanali, A, Zhao, Y, Vedadi, M, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Walker, J.R, Bochkarev, A, Hui, R, Brokx, S.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Characterization of 14-3-3 proteins from Cryptosporidium parvum.
Plos One, 6, 2011
|
|
5T7N
 
 | | X-ray crystal structure of AA13 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, ZINC ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1BTR
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1993-05-25 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
5V2V
 
 | | Ethylene forming enzyme in complex with nickel | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, NICKEL (II) ION | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
5V2Y
 
 | | Ethylene forming enzyme in complex with manganese, 2-oxoglutarate and L-arginine | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.428 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
5IL9
 
 | | Crystal structure of Deg9 | | Descriptor: | GLYCEROL, Protease Do-like 9 | | Authors: | Ouyang, M, Liu, L, Li, X.Y, Zhao, S, Zhang, L.X. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Deg9 reveals a novel octameric-type HtrA protease
Nat Plants, 3, 2017
|
|
8EDP
 
 | | Crystal structure of a three-tetrad, parallel, and K+ stabilized homopurine G-quadruplex from human chromosome 7 | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*AP*GP*GP*GP*AP*AP*AP*AP*GP*GP*GP*GP*AP*GP*GP*GP*AP*AP*AP*AP*GP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, ... | | Authors: | Ye, M, Yatsunyk, L.A. | | Deposit date: | 2022-09-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Homopurine guanine-rich sequences in complex with N-methyl mesoporphyrin IX form parallel G-quadruplex dimers and display a unique symmetry tetrad.
Bioorg.Med.Chem., 77, 2022
|
|
1BV2
 
 | | LIPID TRANSFER PROTEIN FROM RICE SEEDS, NMR, 14 STRUCTURES | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Poznanski, J, Sodano, P, Suh, S.W, Lee, J.Y, Ptak, M, Vovelle, F. | | Deposit date: | 1998-09-21 | | Release date: | 1999-05-18 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a lipid transfer protein extracted from rice seeds. Comparison with homologous proteins.
Eur.J.Biochem., 259, 1999
|
|
5GQP
 
 | | Thaumatin Structure at pH 8.0, orthorhombic type1 | | Descriptor: | Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | Thaumatin Structure at pH 8.0, orthorhombic type1
To Be Published
|
|
1BTB
 
 | |
5IM1
 
 | |
5GAG
 
 | | RNC in complex with SRP-SR in the closed state | | Descriptor: | 1A9L SS, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Boehringer, D, Leibundgut, M, Ban, N. | | Deposit date: | 2015-11-26 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon.
Nat Commun, 7, 2016
|
|
1BM0
 
 | | CRYSTAL STRUCTURE OF HUMAN SERUM ALBUMIN | | Descriptor: | SERUM ALBUMIN | | Authors: | Sugio, S, Kashima, A, Mochizuki, S, Noda, M, Kobayashi, K. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human serum albumin at 2.5 A resolution.
Protein Eng., 12, 1999
|
|
