3APS
 
 | | Crystal structure of Trx4 domain of ERdj5 | | Descriptor: | DnaJ homolog subfamily C member 10, GLYCEROL, SULFATE ION | | Authors: | Inaba, K, Suzuki, M, Nagata, K. | | Deposit date: | 2010-10-20 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5.
Mol.Cell, 41, 2011
|
|
1KEN
 
 | | INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH AN ANTIBODY THAT PREVENTS THE HEMAGGLUTININ LOW PH FUSOGENIC TRANSITION | | Descriptor: | alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin HA1, hemagglutinin HA2, ... | | Authors: | Barbey-Martin, C, Gigant, B, Bizebard, T, Calder, L.J, Wharto, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 2001-11-16 | | Release date: | 2002-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | An Antibody that Prevents the Hemagglutinin Low pH Fusogenic Transition
VIROLOGY, 294, 2002
|
|
5HNF
 
 | | Crystal structure of pyrene- and phenanthrene-modified DNA in complex with the BpuJ1 endonuclease binding domain | | Descriptor: | DNA (5'-D(*GP*(YPE)P*AP*CP*CP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*CP*GP*GP*GP*TP*(YPF)P*C)-3'), Restriction endonuclease R.BpuJI | | Authors: | Probst, M, Aeschimann, W, Chau, T.-T.-H, Langenegger, S.M, Stocker, A, Haener, R. | | Deposit date: | 2016-01-18 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Structural insight into DNA-assembled oligochromophores: crystallographic analysis of pyrene- and phenanthrene-modified DNA in complex with BpuJI endonuclease.
Nucleic Acids Res., 44, 2016
|
|
8ONZ
 
 | | Chaetomium thermophilum Methionine Aminopeptidase 2 at the 80S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 60S ribosomal protein L25-like protein, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
6Z61
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-[6-azanyl-8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-[6-(2-azanylethylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]purin-9-yl]-5-(hydroxymethyl)oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2020-05-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | New Chemical Probe Targeting Bacterial NAD Kinase.
Molecules, 25, 2020
|
|
6Z65
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | CITRIC ACID, NAD kinase 1, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-4-azanyl-butanamide | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2020-05-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | New Chemical Probe Targeting Bacterial NAD Kinase.
Molecules, 25, 2020
|
|
8OXF
 
 | |
6Z13
 
 | | VEGF-A 13:107 crystallized with 3C bicyclic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, AMINO GROUP, ... | | Authors: | Gaucher, J.-F, Broussy, S, Reille-Seroussi, M. | | Deposit date: | 2020-05-12 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and ITC Characterization of Peptide-Protein Binding: Thermodynamic Consequences of Cyclization Constraints, a Case Study on Vascular Endothelial Growth Factor Ligands.
Chemistry, 2022
|
|
6Z64
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl-(3-azanylpropyl)amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2020-05-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | New Chemical Probe Targeting Bacterial NAD Kinase.
Molecules, 25, 2020
|
|
7SJZ
 
 | | Crystal structure of aS162A mutant of Co-type nitrile hydratase from Pseudonocardia thermophila | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Ogutu, R.A.M.I, Holz, C.R, Bennett, B, St Maurice, M. | | Deposit date: | 2021-10-19 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Examination of the Catalytic Role of the Axial Cystine Ligand in the Co-Type Nitrile Hydratase from Pseudonocardia thermophila JCM 3095
Catalysts, 11, 2021
|
|
6Z3F
 
 | | VEGF-A 13:107 crystallized with 2C bicyclic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, AMINO GROUP, ... | | Authors: | Gaucher, J.-F, Broussy, S, Reille-Seroussi, M. | | Deposit date: | 2020-05-20 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and ITC Characterization of Peptide-Protein Binding: Thermodynamic Consequences of Cyclization Constraints, a Case Study on Vascular Endothelial Growth Factor Ligands.
Chemistry, 2022
|
|
8OPA
 
 | |
5HPK
 
 | | System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes: NEDD4L and UbV NL.1 | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like, Ubiquitin variant NL.1 | | Authors: | Wu, K.-P, Mukherjee, M, Mercredi, P.Y, Schulman, B.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
8OPE
 
 | |
5HN5
 
 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and isocitrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Homoisocitrate dehydrogenase, ISOCITRIC ACID, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
7RY5
 
 | | Cellular Retinoic Acid Binding Protein II with Bound Inhibitor 4-[6-({4-[(fluorosulfonyl)oxy]phenyl}ethynyl)-4,4-dimethyl-3,4-dihydroquinolin-1(2H)-yl]-4-oxobutanoic acid | | Descriptor: | 4-[6-({4-[(fluorosulfonyl)oxy]phenyl}ethynyl)-4,4-dimethyl-3,4-dihydroquinolin-1(2H)-yl]-4-oxobutanoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Kimmel, B.R, Mrksich, M. | | Deposit date: | 2021-08-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an Enzyme-Inhibitor Reaction Using Cellular Retinoic Acid Binding Protein II for One-Pot Megamolecule Assembly.
Chemistry, 27, 2021
|
|
8OPK
 
 | |
3AQA
 
 | | Crystal structure of the human BRD2 BD1 bromodomain in complex with a BRD2-interactive compound, BIC1 | | Descriptor: | 1-[2-(1H-benzimidazol-2-ylsulfanyl)ethyl]-3-methyl-1,3-dihydro-2H-benzimidazole-2-thione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Umehara, T, Nakamura, Y, Terada, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-10-27 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Real-Time Imaging of Histone H4K12-Specific Acetylation Determines the Modes of Action of Histone Deacetylase and Bromodomain Inhibitors
Chem.Biol., 18, 2011
|
|
8OPF
 
 | |
8OR4
 
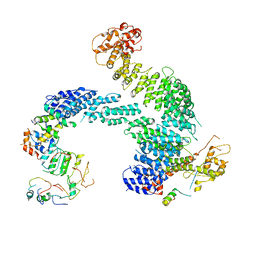 | | Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, Cyclin-dependent kinase 2, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OR3
 
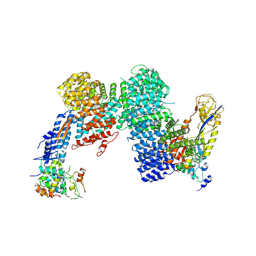 | | CAND1-CUL1-RBX1-SKP1-SKP2-DCNL1 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, DCN1-like protein 1, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
1KNO
 
 | |
8OR2
 
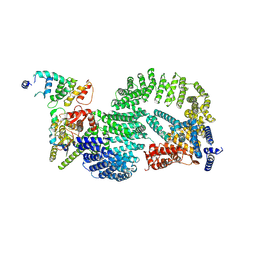 | | CAND1-CUL1-RBX1-DCNL1 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, DCN1-like protein 1, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
5JTA
 
 | |
8OR0
 
 | | CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, Cyclin-dependent kinase 2, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
