5SAV
 
 | | DDR1, N-[2-[3-(2-aminopyrimidin-5-yl)oxyphenyl]ethyl]-3-(trifluoromethoxy)benzamide, 1.760A, P212121, Rfree=23.5% | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-(2-{3-[(2-aminopyrimidin-5-yl)oxy]phenyl}ethyl)-3-(trifluoromethoxy)benzamide | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
5SAY
 
 | | DDR1, N-[2-[3-(2-aminopyrimidin-5-yl)oxyphenyl]ethyl]-3-(trifluoromethoxy)benzamide, 2.190A, P1211, Rfree=27.7% | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-(2-{3-[(2-aminopyrimidin-5-yl)oxy]phenyl}ethyl)-3-(trifluoromethoxy)benzamide | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
5SB2
 
 | | DDR1, 3-chloro-N-[(1R,2S)-2-phenylcyclopropyl]-5-(1H-pyrrolo[2,3-b]pyridin-5-yloxymethyl)benzamide, 1.600A, P212121, Rfree=23.2% | | Descriptor: | 3-chloro-N-[(1R,2S)-2-phenylcyclopropyl]-5-{[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]methyl}benzamide, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Kocer, B, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
5SB1
 
 | | DDR1, 4-chloro-N-[(3S,4R)-4-phenylpyrrolidin-3-yl]-3-(1H-pyrrolo[2,3-b]pyridin-5-yloxymethyl)benzamide, 1.530A, P212121, Rfree=21.4% | | Descriptor: | 4-chloro-N-[(3S,4R)-4-phenylpyrrolidin-3-yl]-3-{[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]methyl}benzamide, Epithelial discoidin domain-containing receptor 1, SULFATE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
5SAZ
 
 | | DDR1, 3-chloro-N-[4-chloro-3-(1H-pyrrolo[2,3-b]pyridin-5-ylcarbamoyl)phenyl]-4-(2-hydroxyethylamino)benzamide, 1.802A, P212121, Rfree=22.2% | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-5-{3-chloro-4-[(2-hydroxyethyl)amino]benzamido}-N-(1H-pyrrolo[2,3-b]pyridin-5-yl)benzamide, CHLORIDE ION, ... | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
5SAW
 
 | | DDR1, 2-[3-(2-pyridin-3-ylethynyl)phenyl]-N-[3-(trifluoromethyl)phenyl]acetamide, 1.601A, P212121, Rfree=22.6% | | Descriptor: | 2-{3-[(pyridin-3-yl)ethynyl]phenyl}-N-[3-(trifluoromethyl)phenyl]acetamide, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
5SAU
 
 | | DDR1, 3-[2-(6-aminopyridin-3-yl)ethynyl]-N-[3-(trifluoromethyl)phenyl]benzamide, 1.800A, P212121, Rfree=23.1% | | Descriptor: | 3-[(6-aminopyridin-3-yl)ethynyl]-N-[3-(trifluoromethyl)phenyl]benzamide, Epithelial discoidin domain-containing receptor 1, MALONATE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
6AKK
 
 | | Crystal structure of the second Coiled-coil domain of SIKE1 | | Descriptor: | GLYCEROL, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
5SAX
 
 | | DDR1, 2-[3-(2-pyridin-3-ylethynyl)phenyl]-N-[3-(trifluoromethyl)phenyl]acetamide, 1.902A, second P212121 form, Rfree=25.4%, second form | | Descriptor: | 2-{3-[(pyridin-3-yl)ethynyl]phenyl}-N-[3-(trifluoromethyl)phenyl]acetamide, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
5SB0
 
 | | DDR1, N-[[2-(2-pyridin-3-yloxyethyl)cyclohexyl]methyl]-3-(trifluoromethoxy)benzamide, 1.970A, P212121, Rfree=25.6% | | Descriptor: | CHLORIDE ION, Epithelial discoidin domain-containing receptor 1, IODIDE ION, ... | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
6AJB
 
 | | Crystal structure of Trypanosoma brucei glycosomal isocitrate dehydrogenase in complex with NADH, alpha-ketoglutarate and ca2+ | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, Isocitrate dehydrogenase [NADP], ... | | Authors: | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
6AKL
 
 | | Crystal structure of Striatin3 in complex with SIKE1 Coiled-coil domain | | Descriptor: | Striatin-3, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
6AL7
 
 | | Crystal structure HpiC1 F138S | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5OTO
 
 | | The crystal structure of CK2alpha in complex with compound 30 | | Descriptor: | 2-(5-chloranyl-1~{H}-benzimidazol-2-yl)-~{N}-[[3-chloranyl-4-(2-ethylphenyl)phenyl]methyl]ethanamine, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
1X3E
 
 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium smegmatis | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2005-05-04 | | Release date: | 2005-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2LQV
 
 | | YebF | | Descriptor: | Protein yebF | | Authors: | Prehna, G, Zhang, G, Gong, X, Duszyk, M, Okon, M, Mcintosh, L.P, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2012-03-16 | | Release date: | 2012-06-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Protein Export Pathway Involving Escherichia coli Porins.
Structure, 20, 2012
|
|
2LRI
 
 | | NMR structure of the second PHD finger of AIRE (AIRE-PHD2) | | Descriptor: | Autoimmune regulator, ZINC ION | | Authors: | Gaetani, M, Chignola, F, Mollica, L, Quilici, G, Mannella, V, Spiliotopoulos, D, Musco, G. | | Deposit date: | 2012-04-03 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | AIRE-PHD fingers are structural hubs to maintain the integrity of chromatin-associated interactome.
Nucleic Acids Res., 40, 2012
|
|
2LMR
 
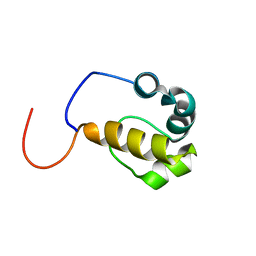 | | Solution structure of the first sam domain of odin | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 1A | | Authors: | Leone, M, Mercurio, F. | | Deposit date: | 2011-12-12 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the First Sam Domain of Odin and Binding Studies with the EphA2 Receptor.
Biochemistry, 51, 2012
|
|
6AA8
 
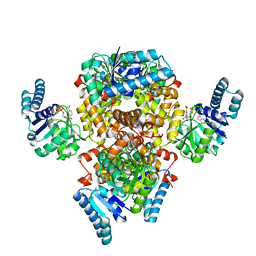 | |
4ZRA
 
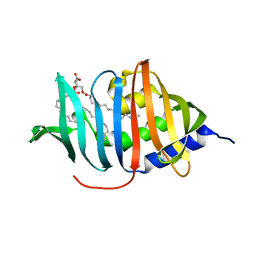 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS LPRG BINDING TO TRIACYLGLYCERIDE | | Descriptor: | Lipoprotein LprG, Tripalmitoylglycerol | | Authors: | Martinot, A.J, Farrow, M, Bai, L, Layre, E, Cheng, T.Y, Tsai, J.H.C, Iqbal, J, Annand, J, Sullivan, Z, Hussain, M, Sacchettini, J, Moody, D.B, Seeliger, J, Rubin, E.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-05-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium tuberculosis.
Plos Pathog., 12, 2016
|
|
6A5I
 
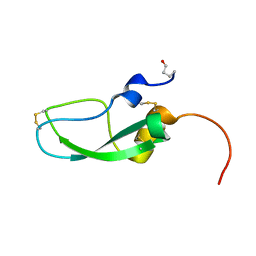 | | Pseudocerastes Persicus Trypsin Inhibitor | | Descriptor: | Trypsin Inhibitor | | Authors: | Amininasab, M. | | Deposit date: | 2018-06-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of PPTI, a kunitz-type protein from the venom of Pseudocerastes persicus.
PLoS ONE, 14, 2019
|
|
6ABA
 
 | | The crystal structure of the photoactivated state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven marine bacterial chloride importer in the rhodopsin family
To Be Published
|
|
1IPD
 
 | | THREE-DIMENSIONAL STRUCTURE OF A HIGHLY THERMOSTABLE ENZYME, 3-ISOPROPYLMALATE DEHYDROGENASE OF THERMUS THERMOPHILUS AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | Authors: | Imada, K, Sato, M, Tanaka, N, Katsube, Y, Matsuura, Y, Oshima, T. | | Deposit date: | 1992-01-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution.
J.Mol.Biol., 222, 1991
|
|
6ACQ
 
 | |
2LUB
 
 | |
