4KAW
 
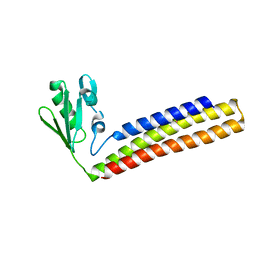 | | Crystal structure of ribosome recycling factor mutant R39G from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-23 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
2OXC
 
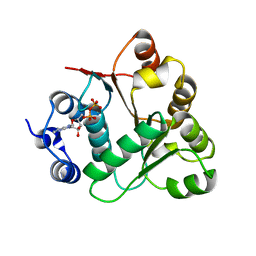 | | Human DEAD-box RNA helicase DDX20, DEAD domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable ATP-dependent RNA helicase DDX20 | | Authors: | Karlberg, T, Ogg, D, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Lehtio, L, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-20 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human DEAD-box RNA helicase DDX20
To be Published
|
|
4KB2
 
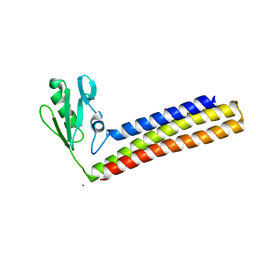 | | Crystal structure of ribosome recycling factor mutant R109A from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-23 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
4KC6
 
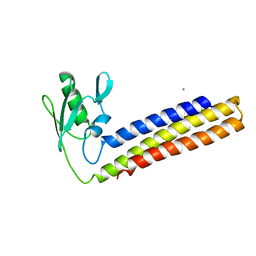 | | Crystal structure of C-terminal deletion mutant of ribosome recycling factor from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-24 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
5BCA
 
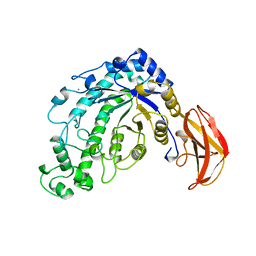 | | BETA-AMYLASE FROM BACILLUS CEREUS VAR. MYCOIDES | | Descriptor: | CALCIUM ION, PROTEIN (1,4-ALPHA-D-GLUCAN MALTOHYDROLASE.) | | Authors: | Oyama, T, Kusunoki, M, Kishimoto, Y, Takasaki, Y, Nitta, Y. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-amylase from Bacillus cereus var. mycoides at 2.2 A resolution.
J.Biochem.(Tokyo), 125, 1999
|
|
7KNG
 
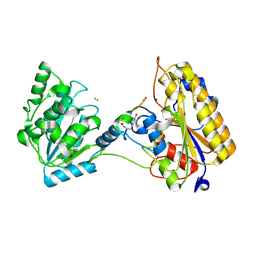 | | 2.10A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-2 Y7F) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, DTY-ASP-TYR-PRO-GLY-ASP-PHE-CYS-TYR-LEU-TYR-GLY-THR-CYS, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
7KNF
 
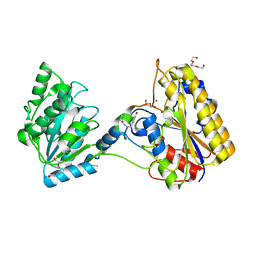 | | 1.80A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-1 NHOH) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, DTY-ASP-TYR-PRO-GLY-ASP-HIS-CYS-TYR-LEU-TYR-GLY-THR, SODIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
1V8Z
 
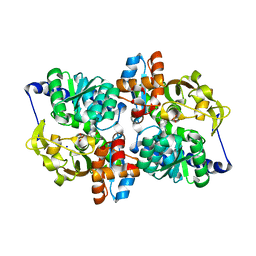 | | X-ray crystal structure of the Tryptophan Synthase b2 Subunit from Hyperthermophile, Pyrococcus furiosus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Hioki, Y, Ogasahara, K, Lee, S.J, Ma, J, Ishida, M, Yamagata, Y, Matsuura, Y, Ota, M, Kuramitsu, S, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of the tryptophan synthase beta subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors
Eur.J.Biochem., 271, 2004
|
|
5VG2
 
 | | Intradiol ring-cleavage Dioxygenase from Tetranychus urticae | | Descriptor: | CACODYLATE ION, FE (III) ION, Intradiol ring-cleavage Dioxygenase | | Authors: | Schlachter, C, Klapper, V, Chruszcz, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and functional characterization of an intradiol ring-cleavage dioxygenase from the polyphagous spider mite herbivore Tetranychus urticae Koch.
Insect Biochem.Mol.Biol., 107, 2019
|
|
3O57
 
 | | Catalytic domain of human phosphodiesterase 4b2b in complex with a 5-heterocycle pyrazolopyridine inhibitor | | Descriptor: | 5-[5-benzyl-4-(2-oxo-2-pyrrolidin-1-ylethyl)-1,3-oxazol-2-yl]-1-ethyl-N-(tetrahydro-2H-pyran-4-yl)-1H-pyrazolo[3,4-b]pyridin-4-amine, ARSENIC, GLYCEROL, ... | | Authors: | Somers, D.O, Neu, M. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyrazolopyridines as potent PDE4B inhibitors: 5-heterocycle SAR.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O5U
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with protocatechuate | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
5VIM
 
 | |
1BJN
 
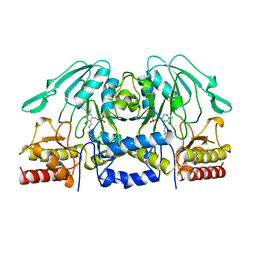 | |
5S1W
 
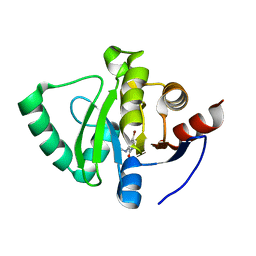 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z838838708 | | Descriptor: | N-(5-bromo-2-oxo-1,2-dihydropyridin-3-yl)acetamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.135 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5SAF
 
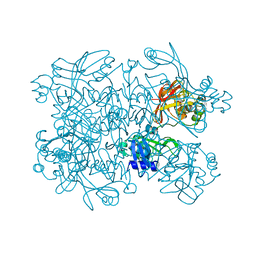 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-321461 | | Descriptor: | 6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S2F
 
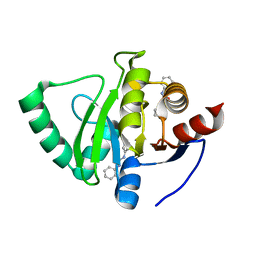 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z44592329 | | Descriptor: | N-phenyl-N'-pyridin-3-ylurea, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.186 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S2U
 
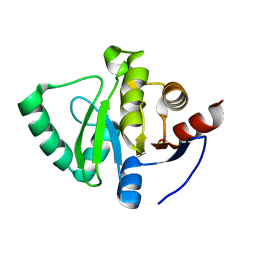 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z85956652 | | Descriptor: | N-(3-chloro-2-methylphenyl)glycinamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.034 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3A
 
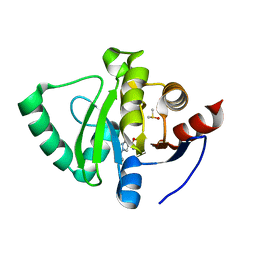 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1562205518 | | Descriptor: | 1-(2-hydroxyethyl)-1H-pyrazole-4-carboxamide, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.178 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
6K4G
 
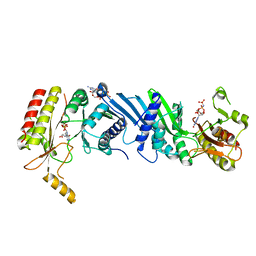 | | Crystal structure of the PI5P4Kbeta-GMPPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Senda, M, Senda, T. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 2022
|
|
5S3G
 
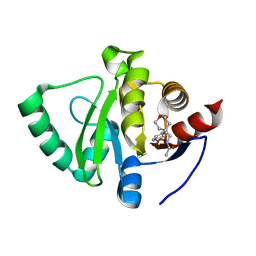 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z384468096 | | Descriptor: | N-[(4-phenyloxan-4-yl)methyl]acetamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3X
 
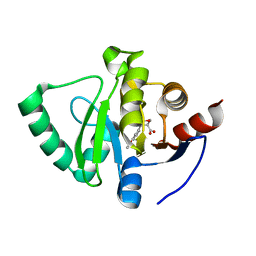 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with POB0136 | | Descriptor: | (3S,4S)-4-(3-methoxyphenyl)oxane-3-carboxylic acid, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1ULQ
 
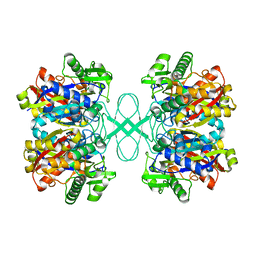 | | Crystal structure of tt0182 from Thermus thermophilus HB8 | | Descriptor: | putative acetyl-CoA acetyltransferase | | Authors: | Ago, H, Hamada, K, Ida, K, Kanda, H, Sugahara, M, Yamamoto, M, Kuroishi, C, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of tt0182 from Thermus thermophilus HB8
To be Published
|
|
5S4E
 
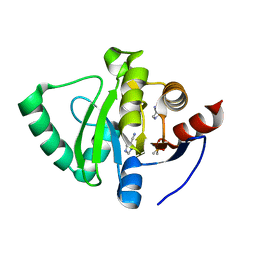 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z2301685688 | | Descriptor: | 1H-imidazole-5-carbonitrile, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3OVZ
 
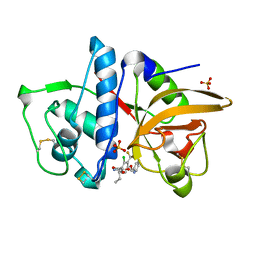 | | Cathepsin K in complex with a covalent inhibitor with a ketoamide warhead | | Descriptor: | Cathepsin K, N-[(1S)-3-amino-1-ethyl-2,3-dioxopropyl]-2-chloro-4-(pyridin-2-ylmethoxy)-3-(trifluoromethyl)benzamide, SULFATE ION | | Authors: | Fradera, X, van Zeeland, M, Uitdehaag, J.C.M. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Trifluoromethylphenyl as P2 for ketoamide-based cathepsin S inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5SAI
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z1424343998 | | Descriptor: | N-{2-[(propan-2-yl)sulfanyl]phenyl}urea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
