8QF0
 
 | |
2P05
 
 | | Structural Insights into the Evolution of a Non-Biological Protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Smith, M, Rosenow, M, Wang, M, Allen, J.P, Szostak, J.W, Chaput, J.C. | | Deposit date: | 2007-02-28 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the evolution of a non-biological protein: importance of surface residues in protein fold optimization.
PLoS ONE, 2, 2007
|
|
4Q7G
 
 | | 1.7 Angstrom Crystal Structure of leukotoxin LukD from Staphylococcus aureus. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Leucotoxin LukDv | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Shatsman, S, Kwon, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the components of the Staphylococcus aureus leukotoxin ED.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6Z19
 
 | | Crystal structure of P8C9 bound to CK2alpha | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2020-05-13 | | Release date: | 2021-05-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
2PCA
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION | | Authors: | Sugahara, M, Taketa, M, Morikawa, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
1PRB
 
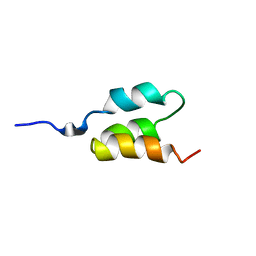 | | STRUCTURE OF AN ALBUMIN-BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN PAB | | Authors: | Johansson, M.U, De Chateau, M, Wikstrom, M, Forsen, S, Drakenberg, T, Bjorck, L. | | Deposit date: | 1997-01-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the albumin-binding GA module: a versatile bacterial protein domain.
J.Mol.Biol., 266, 1997
|
|
6WXC
 
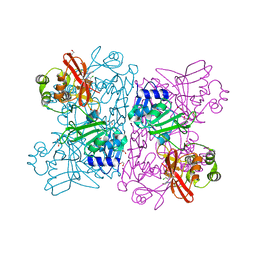 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with potential repurposing drug Tipiracil | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL, FORMIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-10 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6U0S
 
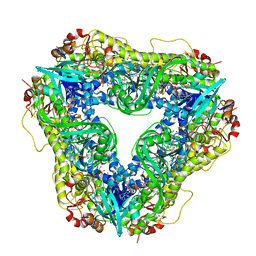 | | Crystal structure of the flavin-dependent monooxygenase PieE in complex with FAD and substrate | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, 2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraen-1-yl]-6-methoxy-3-methylpyridin-4-ol, CHLORIDE ION, ... | | Authors: | Shi, R, Manenda, M. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
5CHS
 
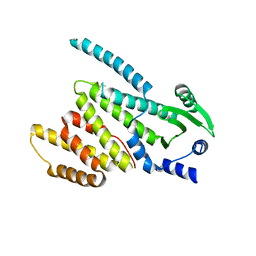 | |
1PIH
 
 | | THE THREE DIMENSIONAL STRUCTURE OF THE PARAMAGNETIC PROTEIN HIPIP I FROM E.HALOPHILA THROUGH NUCLEAR MAGNETIC RESONANCE | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Eltis, L.D, Felli, I, Kastrau, D.H.W, Luchinat, C, Piccioli, M, Pierattelli, R, Smith, M. | | Deposit date: | 1994-08-03 | | Release date: | 1994-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure in solution of the paramagnetic high-potential iron-sulfur protein I from Ectothiorhodospira halophila through nuclear magnetic resonance.
Eur.J.Biochem., 225, 1994
|
|
1PIJ
 
 | | THE THREE DIMENSIONAL STRUCTURE OF THE PARAMAGNETIC PROTEIN HIPIP I FROM E.HALOPHILA THROUGH NUCLEAR MAGNETIC RESONANCE | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Eltis, L.D, Felli, I.C, Kastrau, D.H.W, Luchinat, C, Piccioli, M, Pierattelli, R, Smith, M. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure in solution of the paramagnetic high-potential iron-sulfur protein I from Ectothiorhodospira halophila through nuclear magnetic resonance.
Eur.J.Biochem., 225, 1994
|
|
5OVN
 
 | | Crystal Structure of FIV Reverse Transcriptase | | Descriptor: | POL protein | | Authors: | Galilee, M, Alian, A. | | Deposit date: | 2017-08-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.942 Å) | | Cite: | The structure of FIV reverse transcriptase and its implications for non-nucleoside inhibitor resistance.
PLoS Pathog., 14, 2018
|
|
7PZZ
 
 | | Crystal structure of serine hydroxymethyltransferase, isoform 2 from Arabidopsis thaliana (SHM2) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Ruszkowski, M, Sekula, B. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
5OX8
 
 | | Structure of Enhanced Cyan Fluorescent Protein at pH 5.0 | | Descriptor: | CHLORIDE ION, Green fluorescent protein | | Authors: | Gotthard, G, von Stetten, D, Clavel, D, Noirclerc-Savoye, M, Royant, A. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Chromophore Isomer Stabilization Is Critical to the Efficient Fluorescence of Cyan Fluorescent Proteins.
Biochemistry, 56, 2017
|
|
1ZN1
 
 | | Coordinates of RRF fitted into Cryo-EM map of the 70S post-termination complex | | Descriptor: | 30S ribosomal protein S12, Ribosome recycling factor, ribosomal 16S RNA, ... | | Authors: | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
6R7U
 
 | | Selenomethionine variant of Tannerella forsythia promirolysin mutant E225A | | Descriptor: | BORIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Rodriguez-Banqueri, A, Guevara, T, Ksiazek, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2019-03-29 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based mechanism of cysteine-switch latency and of catalysis by pappalysin-family metallopeptidases.
Iucrj, 7, 2020
|
|
5OYH
 
 | | crystal structure of the catalytic core of a rhodopsin-guanylyl cyclase with converted specificity in complex with ATPalphaS | | Descriptor: | ADENOSINE-5'-SP-ALPHA-THIO-TRIPHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Broser, M, Scheib, U, Hegemann, P. | | Deposit date: | 2017-09-09 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Rhodopsin-cyclases for photocontrol of cGMP/cAMP and 2.3 angstrom structure of the adenylyl cyclase domain.
Nat Commun, 9, 2018
|
|
6R35
 
 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PAO1 in complex with lewis x tetrasaccharide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Lepsik, M, Sommer, R, Kuhaudomlarp, S, Lelimousin, M, Varrot, A, Titz, A, Imberty, A. | | Deposit date: | 2019-03-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Induction of rare conformation of oligosaccharide by binding to calcium-dependent bacterial lectin: X-ray crystallography and modelling study.
Eur.J.Med.Chem., 177, 2019
|
|
7Q00
 
 | | Crystal structure of serine hydroxymethyltransferase, isoform 4 from Arabidopsis thaliana (SHM4) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Serine hydroxymethyltransferase 4 | | Authors: | Ruszkowski, M, Sekula, B. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
6WLC
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Chang, C, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-19 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6U0P
 
 | | Crystal structure of PieE, the flavin-dependent monooxygenase involved in the biosynthesis of piericidin A1 | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Shi, R, Manenda, M, Picard, M.-E. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
5FP5
 
 | | Structure of cyclin-dependent kinase 2 with small-molecule ligand 4- fluorobenzoic acid (AT222) in an alternate binding site. | | Descriptor: | 4-fluorobenzoic acid, ACETYL GROUP, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Jhoti, H, Ludlow, R.F, O'Reilly, M, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4MFI
 
 | | Crystal structure of Mycobacterium tuberculosis UgpB | | Descriptor: | Sn-glycerol-3-phosphate ABC transporter substrate-binding protein UspB | | Authors: | Jiang, D, Bartlam, M, Rao, Z. | | Deposit date: | 2013-08-28 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of Mycobacterium tuberculosis ATP-binding cassette transporter subunit UgpB reveals specificity for glycerophosphocholine
Febs J., 281, 2014
|
|
5JD5
 
 | | Crystal structure of MGS-MilE3, an alpha/beta hydrolase enzyme from the metagenome of pyrene-phenanthrene enrichment culture with sediment sample of Milazzo Harbor, Italy | | Descriptor: | CHLORIDE ION, MGS-MilE3 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Martinez-Martinez, M, Chernikova, T.N, Golyshin, P.N, Yakimov, M.M, Ferrer, M, Savchenko, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of MGS-MilE3, an alpha/beta hydrolase enzyme from the metagenome of pyrene-phenanthrene enrichment culture with sediment sample of Milazzo Harbor, Italy
To Be Published
|
|
6R3B
 
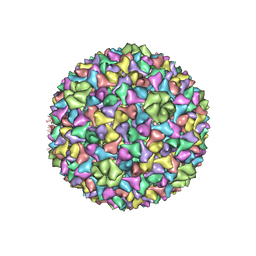 | | BACTERIOPHAGE SPP1 PROCAPSID-I PROTEIN | | Descriptor: | Major capsid protein | | Authors: | Ignatiou, A, Brasiles, S, El Sadek-Fadel, M, Buerger, J, Mielke, T, Topf, M, Tavares, P. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural transitions during the scaffolding-driven assembly of a viral capsid.
Nat Commun, 10, 2019
|
|
