6TJX
 
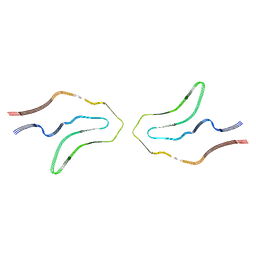 | | Cryo-EM structure of TypeII tau filaments extracted from the brains of individuals with Corticobasal degeneration | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Murzin, A.G, Falcon, B, Shi, Y, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Novel tau filament fold in corticobasal degeneration.
Nature, 580, 2020
|
|
5H15
 
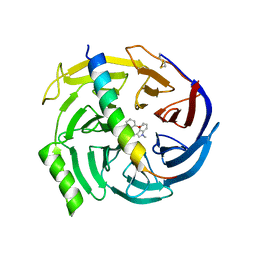 | | EED in complex with PRC2 allosteric inhibitor EED709 | | Descriptor: | (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5O2Y
 
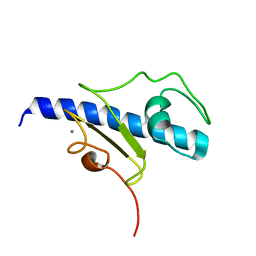 | | NMR structure of the calcium bound form of PulG, major pseudopilin from Klebsiella oxytoca T2SS | | Descriptor: | CALCIUM ION, General secretion pathway protein G | | Authors: | Lopez-Castilla, A, Bardiaux, B, Vitorge, B, Thomassin, J.-L, Zheng, W, Yu, X, Egelman, E.H, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2017-05-23 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the calcium-dependent type 2 secretion pseudopilus.
Nat Microbiol, 2, 2017
|
|
2CMB
 
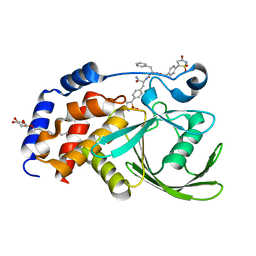 | | Structural Basis for Inhibition of Protein Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics | | Descriptor: | N-{[4-(1,1-DIOXIDO-3-OXO-2,3-DIHYDROISOTHIAZOL-5-YL)PHENYL]ACETYL}-L-PHENYLALANYL-4-(1,1-DIOXIDO-3-OXO-2,3-DIHYDROISOTHIAZOL-5-YL)-L-PHENYLALANINAMIDE, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1, octyl beta-D-glucopyranoside | | Authors: | Ala, P.J, Gonneville, L, Hillman, M.C, Becker-Pasha, M, Wei, M, Reid, B.G, Klabe, R, Yue, E.W, Wayland, B, Douty, B, Combs, A.P, Polam, P, Wasserman, Z, Bower, M, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-04 | | Release date: | 2006-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Inhibition of Protein-Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics.
J.Biol.Chem., 281, 2006
|
|
2CKK
 
 | | High resolution crystal structure of the human kin17 C-terminal domain containing a kow motif | | Descriptor: | ACETATE ION, IODIDE ION, KIN17 | | Authors: | le Maire, A, Schiltz, M, Pinon-Lataillade, G, Stura, E, Couprie, J, Gondry, M, Angulo-Mora, J, Zinn-Justin, S. | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Tandem of SH3-Like Domains Participates in RNA Binding in Kin17, a Human Protein Activated in Response to Genotoxics.
J.Mol.Biol., 364, 2006
|
|
7B61
 
 | | Crystal structure of MurE from E.coli in complex with Z57299526 | | Descriptor: | (R)-N-(1-cyclopropylethyl)-6-methylpicolinamide, (S)-N-(1-cyclopropylethyl)-6-methylpicolinamide, CITRIC ACID, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
5G5X
 
 | |
7B6O
 
 | | Crystal structure of E.coli MurE mutant - C269S C340S C450S | | Descriptor: | ISOPROPYL ALCOHOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
6GVK
 
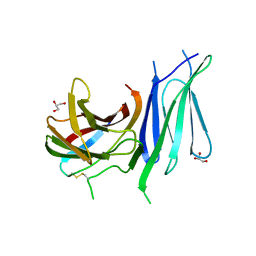 | | Second pair of Fibronectin type III domains of integrin beta4 (T1663R mutant) bound to the bullous pemphigoid antigen BP230 (BPAG1e) | | Descriptor: | Dystonin, GLYCEROL, Integrin beta-4 | | Authors: | Manso, J.A, Gomez-Hernandez, M, Alonso-Garcia, N, de Pereda, J.M. | | Deposit date: | 2018-06-21 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Integrin alpha 6 beta 4 Recognition of a Linear Motif of Bullous Pemphigoid Antigen BP230 Controls Its Recruitment to Hemidesmosomes.
Structure, 27, 2019
|
|
6GVS
 
 | | Engineered glycolyl-CoA reductase comprising 8 mutations with bound NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION | | Authors: | Zarzycki, J, Trudeau, D, Scheffen, M, Erb, T.J, Tawfik, D.S. | | Deposit date: | 2018-06-21 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.579 Å) | | Cite: | Design and in vitro realization of carbon-conserving photorespiration.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6TRR
 
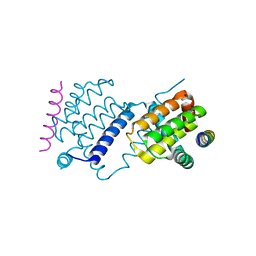 | |
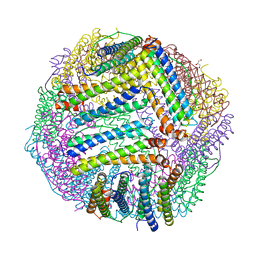
5JKL
 
 | |
7B6K
 
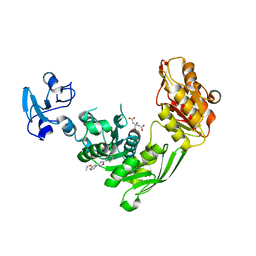 | | Crystal structure of MurE from E.coli in complex with Z57715447 | | Descriptor: | 5-cyclohexyl-3-(pyridin-4-yl)-1,2,4-oxadiazole, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
6TRI
 
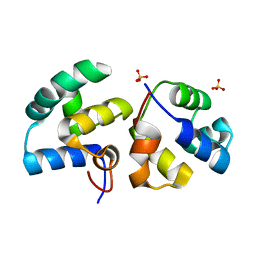 | | CI-MOR repressor-antirepressor complex of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | CI, MOR, SULFATE ION | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Jensen, M.R, Lo Leggio, L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2PQ4
 
 | |
6TS6
 
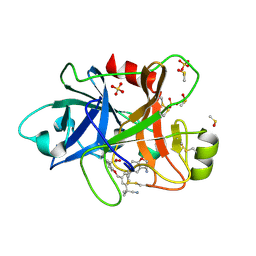 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-[(3~{S})-3-azanyl-2,3-dihydro-1-benzofuran-5-yl]-5-(2-cyanopropan-2-yl)phenyl]methoxy]phenyl]ethanoic acid, Coagulation factor XI, DIMETHYL SULFOXIDE, ... | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6GQE
 
 | |
1XSO
 
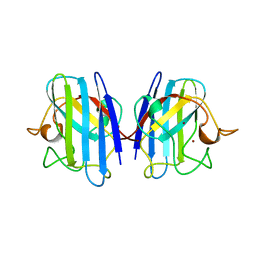 | | THREE-DIMENSIONAL STRUCTURE OF XENOPUS LAEVIS CU,ZN SUPEROXIDE DISMUTASE B DETERMINED BY X-RAY CRYSTALLOGRAPHY AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Djinovic Carugo, K, Coda, A, Battistoni, A, Carri, M.T, Polticelli, F, Desideri, A, Rotilio, G, Wilson, K.S, Bolognesi, M. | | Deposit date: | 1995-03-14 | | Release date: | 1995-07-10 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Three-dimensional structure of Xenopus laevis Cu,Zn superoxide dismutase b determined by X-ray crystallography at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
3NHP
 
 | |
7B6L
 
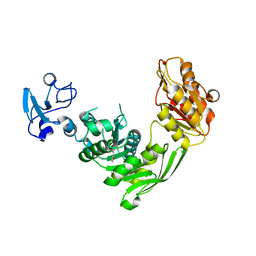 | | Crystal structure of MurE from E.coli in complex with Z57299368 | | Descriptor: | (1-ethyl-1H-benzoimidazol-2-yl)-furan-2-ylmethyl-aminee, ISOPROPYL ALCOHOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
3N5K
 
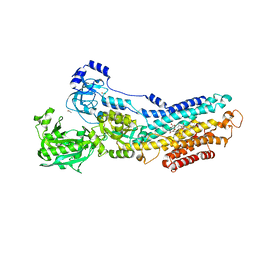 | | Structure Of The (Sr)Ca2+-ATPase E2-AlF4- Form | | Descriptor: | ACETATE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Bublitz, M, Olesen, C, Poulsen, H, Morth, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2010-05-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ion pathways in the sarcoplasmic reticulum Ca2+-ATPase.
J.Biol.Chem., 288, 2013
|
|
2V8C
 
 | | Mouse Profilin IIa in complex with the proline-rich domain of VASP | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, PROFILIN-2, ... | | Authors: | Kursula, P, Downer, J, Witke, W, Wilmanns, M. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High-Resolution Structural Analysis of Mammalian Profilin 2A Complex Formation with Two Physiological Ligands: The Formin Homology 1 Domain of Mdia1 and the Proline-Rich Domain of Vasp.
J.Mol.Biol., 375, 2008
|
|
1V7X
 
 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
6IMV
 
 | | The complex structure of endo-beta-1,2-glucanase from Talaromyces funiculosus with sophorose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanaka, N, Nakajima, M, Narukawa-Nara, M, Matsunaga, H, Kamisuki, S, Aramasa, H, Takahashi, Y, Sugimoto, N, Abe, K, Miyanaga, A, Yamashita, T, Sugawara, F, Kamakura, T, Komba, S, Nakai, H, Taguchi, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification, characterization, and structural analyses of a fungal endo-beta-1,2-glucanase reveal a new glycoside hydrolase family.
J.Biol.Chem., 294, 2019
|
|
6UMW
 
 | | Crystal structure of hEphB1 bound with chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, Ephrin type-B receptor 1 | | Authors: | Ahmed, M, Wang, P, Sadek, H. | | Deposit date: | 2019-10-10 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
