5OD7
 
 | | Hsp90 inhibitor desolvation as a rationale to steer on-rates and impact residence time | | Descriptor: | Heat shock protein HSP 90-alpha, [2-azanyl-6-[2-(4-methylpiperazin-1-yl)sulfonylphenyl]quinazolin-4-yl]-(1,3-dihydroisoindol-2-yl)methanone | | Authors: | Schuetz, D.A, Richter, L, Amaral, M, Grandits, M, Musil, D, Graedler, U, Buchstaller, H.-P, Eggenweiler, H.-M, Frech, M, Ecker, G.F, Lehmann, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Desolvation Steers On-Rate and Impacts Drug Residence Time of Heat Shock Protein 90 (Hsp90) Inhibitors.
J.Med.Chem., 61, 2018
|
|
1S56
 
 | | Crystal Structure of "Truncated" Hemoglobin N (HbN) from Mycobacterium tuberculosis, Soaked with Xe Atoms | | Descriptor: | CYANIDE ION, HEME C, Hemoglobin-like protein HbN, ... | | Authors: | Milani, M, Pesce, A, Ouellet, Y, Dewilde, S, Friedman, J, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2004-01-20 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Heme-ligand tunneling in group I truncated hemoglobins
J.Biol.Chem., 279, 2004
|
|
4JLG
 
 | | SETD7 in complex with inhibitor (R)-PFI-2 and S-adenosyl-methionine | | Descriptor: | 8-fluoro-N-{(2R)-1-oxo-1-(pyrrolidin-1-yl)-3-[3-(trifluoromethyl)phenyl]propan-2-yl}-1,2,3,4-tetrahydroisoquinoline-6-sulfonamide, Histone-lysine N-methyltransferase SETD7, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Wu, H, Zeng, H, El Bakkouri, M, Barsyte, D, Vedadi, M, Tatlock, J, Owen, D, Bunnage, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1S61
 
 | | Crystal Structure of "Truncated" Hemoglobin N (HbN) from Mycobacterium tuberculosis, Soaked with Butyl-isocyanide | | Descriptor: | CYANIDE ION, Hemoglobin-like protein HbN, N-BUTYL ISOCYANIDE, ... | | Authors: | Milani, M, Pesce, A, Ouellet, Y, Dewilde, S, Friedman, J, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2004-01-22 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Heme-ligand tunneling in group I truncated hemoglobins
J.Biol.Chem., 279, 2004
|
|
6G5T
 
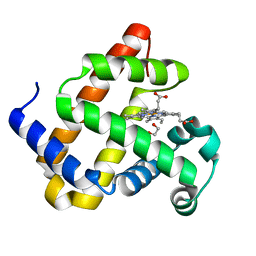 | | Myoglobin H64V/V68A in the resting state, 1.5 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | Deposit date: | 2018-03-30 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
6DWU
 
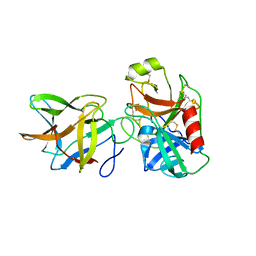 | | Crystal structure of complex of BBKI and Bovine Trypsin | | Descriptor: | Cationic trypsin, Kunitz-type inihibitor | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2018-06-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Crystal structures of the complex of a kallikrein inhibitor from Bauhinia bauhinioides with trypsin and modeling of kallikrein complexes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8OK8
 
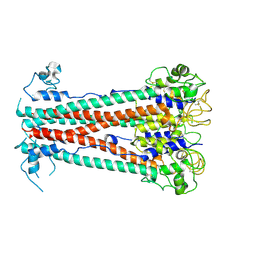 | | Variant Surface Glycoprotein VSG615 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Variant surface glycoprotein 615, ... | | Authors: | Zeelen, J.P, Stebbins, C.E, Chandra, M. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
1SDY
 
 | | STRUCTURE SOLUTION AND MOLECULAR DYNAMICS REFINEMENT OF THE YEAST CU,ZN ENZYME SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Djinovic, K, Gatti, G, Coda, A, Antolini, L, Pelosi, G, Desideri, A, Falconi, M, Marmocchi, F, Rotilio, G, Bolognesi, M. | | Deposit date: | 1991-06-14 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure solution and molecular dynamics refinement of the yeast Cu,Zn enzyme superoxide dismutase.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
5W6P
 
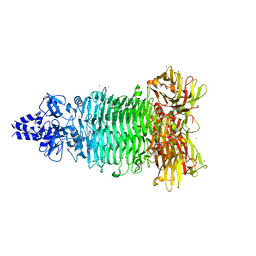 | | Crystal structure of Bacteriophage CBA120 tailspike protein 2 enzymatically active domain (TSP2dN, orf211) | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, ZINC ION, ... | | Authors: | Plattner, M, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2017-06-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.335 Å) | | Cite: | Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120.
J.Mol.Biol., 431, 2019
|
|
4NRT
 
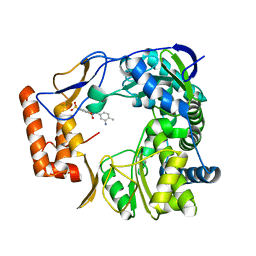 | | Human Norovirus polymerase bound to Compound 6 (suramin derivative) | | Descriptor: | 4-({4-methyl-3-[(3-nitrobenzoyl)amino]benzoyl}amino)naphthalene-1,5-disulfonic acid, hNV-RdRp | | Authors: | Croci, R, Pezzullo, M, Tarantino, D, Mastrangelo, E, Milani, M, Bolognesi, M. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds.
Plos One, 9, 2014
|
|
8OP1
 
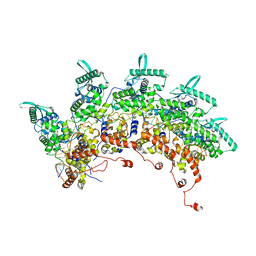 | | Subsection of a helical nucleocapsid of the Respiratory Syncytial Virus | | Descriptor: | Nucleoprotein, RNA (5'-R(P*CP*CP*CP*CP*CP*CP*C)-3') | | Authors: | Gonnin, L, Desfosses, A, Eleouet, J.F, Galloux, M, Gutsche, I. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural landscape of the respiratory syncytial virus nucleocapsids.
Nat Commun, 14, 2023
|
|
8ODN
 
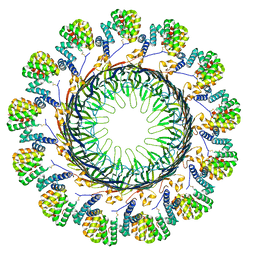 | |
2RTV
 
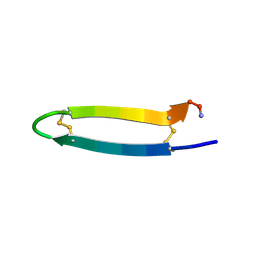 | | Tachyplesin I in water | | Descriptor: | Tachyplesin-1 | | Authors: | Kushibiki, T, Kamiya, M, Aizawa, T, Kumaki, Y, Kikukawa, T, Mizuguchi, M, Demura, M, Kawabata, S.I, Kawano, K. | | Deposit date: | 2013-09-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Interaction between tachyplesin I, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide.
Biochim.Biophys.Acta, 1844, 2014
|
|
7B2L
 
 | | Structure of the endocytic adaptor complex AENTH | | Descriptor: | ANTH domain of Sla2, ENTH domain of epsin Ent1, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Klebl, D.P, Lizarrondo, J, Sobott, F, Garcia-Alai, M, Muench, S.P. | | Deposit date: | 2020-11-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the endocytic adaptor complex reveals the basis for efficient membrane anchoring during clathrin-mediated endocytosis.
Nat Commun, 12, 2021
|
|
6Y35
 
 | | CCAAT-binding complex from Aspergillus fumigatus with cycA DNA | | Descriptor: | CCAAT-binding factor complex subunit HapC, CCAAT-binding factor complex subunit HapE, CCAAT-binding transcription factor subunit HAPB, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2020-02-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of HapE P88L -linked antifungal triazole resistance in Aspergillus fumigatus .
Life Sci Alliance, 3, 2020
|
|
6SS2
 
 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6PD7
 
 | | Crystal Structure of EcDsbA in a complex with purified morpholine carboxylic acid 7 | | Descriptor: | (3R)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-18 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
5O46
 
 | | Crystal structure of Iristatin, a secreted salivary cystatin from the hard tick Ixodes ricinus | | Descriptor: | GLYCEROL, Iristatin | | Authors: | Busa, M, Rezacova, P, Kotal, J, Kotsyfakis, M, Mares, M. | | Deposit date: | 2017-05-26 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The structure and function of Iristatin, a novel immunosuppressive tick salivary cystatin.
Cell.Mol.Life Sci., 76, 2019
|
|
6UWZ
 
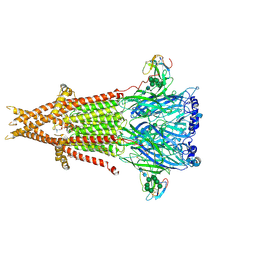 | | Cryo-EM structure of Torpedo acetylcholine receptor in complex with alpha-bungarotoxin | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine receptor subunit alpha, ... | | Authors: | Rahman, M.M, Teng, J, Worrell, B.T, Noveillo, C.M, Lee, M, Karlin, A, Stowell, M, Hibbs, R.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-04-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the Native Muscle-type Nicotinic Receptor and Inhibition by Snake Venom Toxins.
Neuron, 106, 2020
|
|
5HO7
 
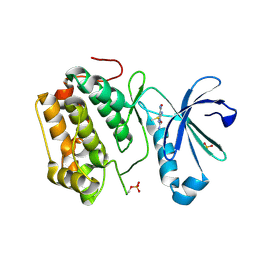 | | DISCOVERY OF NOVEL 7-AZAINDOLES AS PDK1 INHIBITORS | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 5-amino-3-(methylsulfanyl)-1H-pyrazole-1,4-dicarboxamide, SULFATE ION | | Authors: | Wucherer-Plietker, M, Esdar, C, Knoechel, T, Hillertz, P, Heinrich, T, Buchstaller, H.P, Greiner, H, Dorsch, D, Calderini, M, Bruge, D, Mueller, T.J.J, Graedler, U. | | Deposit date: | 2016-01-19 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of novel 7-azaindoles as PDK1 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4JSB
 
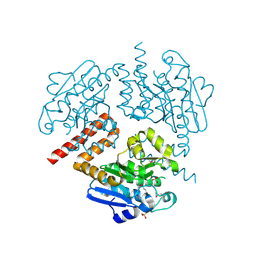 | | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX | | Descriptor: | Enoyl-CoA hydratase, SULFATE ION | | Authors: | Mikolajczak, K, Porebski, P.J, Cooper, D.R, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX
TO BE PUBLISHED
|
|
8OK7
 
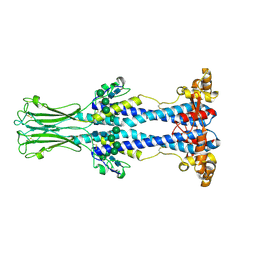 | | Variant Surface Glycoprotein VSG558 NTD | | Descriptor: | Variant surface glycoprotein 558, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zeelen, J.P, Stebbins, C.E, van Straaten, M, Zhong, J. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
5VLK
 
 | |
5VLP
 
 | | PCSK9 complex with LDLR antagonist peptide and Fab7G7 | | Descriptor: | Fab7G7 heavy chain, Fab7G7 light chain, LDLR antagonist peptide, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6FML
 
 | | CryoEM Structure INO80core Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin related protein 5, ... | | Authors: | Eustermann, S, Schall, K, Kostrewa, D, Strauss, M, Hopfner, K. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.
Nature, 556, 2018
|
|
