2GHY
 
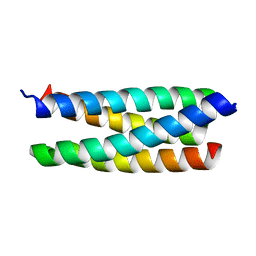 | | Novel Crystal Form of the ColE1 Rom Protein | | Descriptor: | Regulatory protein rop | | Authors: | Jang, S.B, Jeong, M.S, Carter, R.J, Holbrook, E.L, Comolli, L.R, Holbrook, S.R. | | Deposit date: | 2006-03-28 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel crystal form of the ColE1 Rom protein.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1TIJ
 
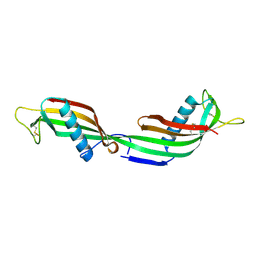 | | 3D Domain-swapped human cystatin C with amyloid-like intermolecular beta-sheets | | Descriptor: | Cystatin C | | Authors: | Janowski, R, Kozak, M, Abrahamson, M, Grubb, A, Jaskolski, M. | | Deposit date: | 2004-06-02 | | Release date: | 2005-07-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | 3D domain-swapped human cystatin C with amyloidlike intermolecular beta-sheets.
Proteins, 61, 2005
|
|
2INN
 
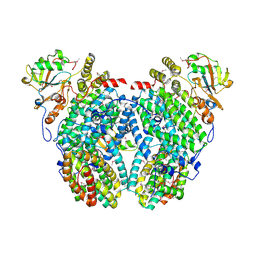 | | Structure of the Phenol Hydroxyalse-Regulatory Protein Complex | | Descriptor: | FE (III) ION, MOLYBDATE ION, Phenol hydroxylase component phL, ... | | Authors: | Sazinsky, M.H, Dunten, P.W, McCormick, M.S, Lippard, S.J. | | Deposit date: | 2006-10-08 | | Release date: | 2007-01-16 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray Structure of a Hydroxylase-Regulatory Protein Complex from a Hydrocarbon-Oxidizing Multicomponent Monooxygenase, Pseudomonas sp. OX1 Phenol Hydroxylase.
Biochemistry, 45, 2006
|
|
2IDY
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Horst, R, Herrmann, T, Joseph, J, Saikatendu, K, Subramanian, V, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2ISA
 
 | | Crystal Structure of Vibrio salmonicida catalase | | Descriptor: | CHLORIDE ION, Catalase, GLYCEROL, ... | | Authors: | Riise, E.K, Lorentzen, M.S, Helland, R, Smalas, A.O, Leiros, H.K.S, Willassen, N.P. | | Deposit date: | 2006-10-17 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The first structure of a cold-active catalase from Vibrio salmonicida at 1.96A reveals structural aspects of cold adaptation
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2ILL
 
 | | Anomalous substructure of Titin-A168169 | | Descriptor: | CHLORIDE ION, Titin | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-10-03 | | Release date: | 2007-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
7L3D
 
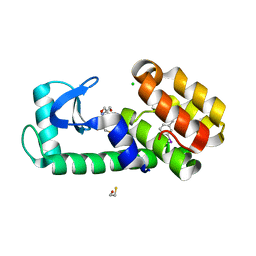 | | T4 Lysozyme L99A - 3-iodotoluene - RT | | Descriptor: | 1-iodo-3-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
8C3W
 
 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ortmayer, M, Levy, C. | | Deposit date: | 2022-12-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
7L3I
 
 | | T4 Lysozyme L99A - propylbenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Endolysin, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
6R21
 
 | | Cryo-EM structure of T7 bacteriophage fiberless tail complex | | Descriptor: | Portal protein, Tail tubular protein gp11, Tail tubular protein gp12 | | Authors: | Cuervo, A, Fabrega-Ferrer, M, Machon, C, Conesa, J.J, Perez-Ruiz, M, Coll, M, Carrascosa, J.L. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
5JMD
 
 | | Heparinase III-BT4657 gene product, Methylated Lysines | | Descriptor: | Heparinase III protein, MAGNESIUM ION | | Authors: | Ulaganathan, T.S, Shi, R, Yao, D, Garron, M.-L, Cherney, M, Cygler, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-05-25 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational flexibility of PL12 family heparinases: structure and substrate specificity of heparinase III from Bacteroides thetaiotaomicron (BT4657).
Glycobiology, 27, 2017
|
|
2P3T
 
 | |
5BY9
 
 | |
6F35
 
 | | Crystal structure of the aspartate aminotranferase from Rhizobium meliloti | | Descriptor: | ACETATE ION, Aspartate aminotransferase B, GLYCEROL, ... | | Authors: | Cobessi, D, Graindorge, M, Giustini, C, Matringe, M. | | Deposit date: | 2017-11-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
6WIQ
 
 | | Crystal structure of the co-factor complex of NSP7 and the C-terminal domain of NSP8 from SARS CoV-2 | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6EXI
 
 | |
5C2T
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with rhodoquinone-2 | | Descriptor: | 2-amino-5-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-3-methoxy-6-methylcyclohexa-2,5-diene-1,4-dione, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-06-16 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
6WGU
 
 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase | | Descriptor: | Corrinoid adenosyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-04-06 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B 12 .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5FOS
 
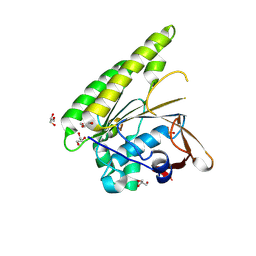 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH OLIGOMERISATION PEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, GLYCEROL, PHOSPHATE ION | | Authors: | Sharpe, T, Moschetti, T, Fischer, G, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
6ESY
 
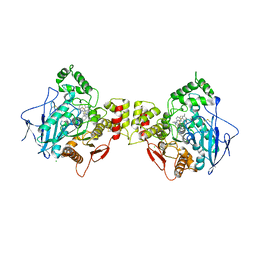 | | Human butyrylcholinesterase in complex with thioflavine T | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nachon, F, Brazzolotto, X, Wandhammer, M, Trovaslet-Leroy, M, Macdonald, I.R, Darvesh, S, Rosenberry, T.L. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparison of the Binding of Reversible Inhibitors to Human Butyrylcholinesterase and Acetylcholinesterase: A Crystallographic, Kinetic and Calorimetric Study.
Molecules, 22, 2017
|
|
1CDZ
 
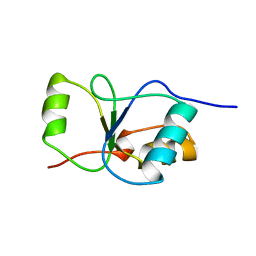 | | BRCT DOMAIN FROM DNA-REPAIR PROTEIN XRCC1 | | Descriptor: | PROTEIN (DNA-REPAIR PROTEIN XRCC1) | | Authors: | Zhang, X, Morera, S, Bates, P, Whitehead, P, Coffer, A, Hainbucher, K, Nash, R, Sternberg, M, Lindahl, T, Freemont, P. | | Deposit date: | 1999-03-04 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an XRCC1 BRCT domain: a new protein-protein interaction module.
EMBO J., 17, 1998
|
|
7SV2
 
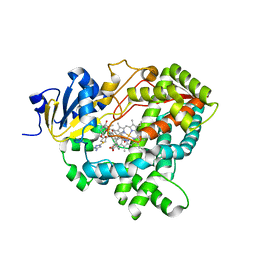 | | Human Cytochrome P450 (CYP) 3A5 ternary complex with azamulin | | Descriptor: | (3aS,4R,5S,6R,8R,9R,9aR,10R)-6-ethyl-5-hydroxy-4,6,9,10-tetramethyl-1-oxodecahydro-3a,9-propanocyclopenta[8]annulen-8-yl [(5-amino-1H-1,2,4-triazol-3-yl)sulfanyl]acetate, Cytochrome P450 3A5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hsu, M, Johnson, E.F. | | Deposit date: | 2021-11-18 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural characterization of the homotropic cooperative binding of azamulin to human cytochrome P450 3A5.
J.Biol.Chem., 298, 2022
|
|
5FV9
 
 | | Crystal structure of GalNAc-T2 in complex with compound 16d | | Descriptor: | 1,2-ETHANEDIOL, GALNAC-T2, GLYCEROL, ... | | Authors: | Ghirardello, M, Rivas, M, Lacetera, A, Delso, I, Lira-Navarrete, E, Tejero, T, Martin-Santamaria, S, Hurtado-Guerrero, R, Merino, P. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Glycomimetics Targeting Glycosyltransferases: Synthetic, Computational and Structural Studies of Less-Polar Conjugates.
Chemistry, 22, 2016
|
|
6R5M
 
 | | Crystal structure of toxin MT9 from mamba venom | | Descriptor: | ACETYL GROUP, Dendroaspis polylepis MT9, GLYCEROL, ... | | Authors: | Stura, E.A, Tepshi, L, Ciolek, J, Triquigneaux, M, Zoukimian, C, De Waard, M, Beroud, R, Servent, D, Gilles, N, Legrand, P, Ciccone, L. | | Deposit date: | 2019-03-25 | | Release date: | 2020-02-12 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MT9, a natural peptide from black mamba venom antagonizes the muscarinic type 2 receptor and reverses the M2R-agonist-induced relaxation in rat and human arteries
Biomed Pharmacother, 150, 2022
|
|
7L5R
 
 | | Crystal Structure of the Oxacillin-hydrolyzing Class D Extended-spectrum Beta-lactamase OXA-14 from Pseudomonas aeruginosa | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Antimicrob.Agents Chemother., 66, 2022
|
|
