8JFC
 
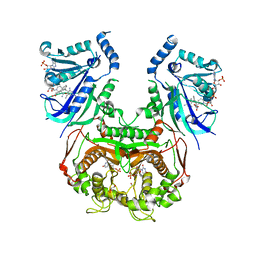 | | V1/S quadruple mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with compound 6 (B21591), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-[3-[[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]methyl]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Saeyang, T, Vitsupakorn, D, Saepua, S, Thongpanchang, C, Yuthavong, Y, Kamchonwongpaisan, S, Hoarau, M. | | Deposit date: | 2023-05-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of rigid biphenyl Plasmodium falciparum DHFR inhibitors using a fragment linking strategy.
Rsc Med Chem, 14, 2023
|
|
8IQK
 
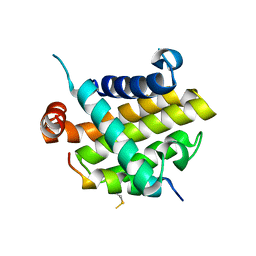 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | Descriptor: | Bcl-2-like protein 1, Bcl-2-modifying factor | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.879 Å) | | Cite: | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
8IQL
 
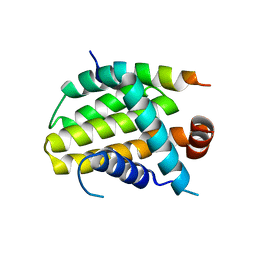 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | Descriptor: | Apoptosis regulator Bcl-2, Bcl-2-modifying factor | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9577 Å) | | Cite: | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
8JFB
 
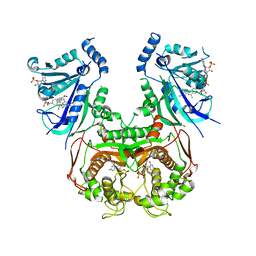 | | V1/S quadruple mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with compound 4 (B21588), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 3-[4-[[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]methyl]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Saeyang, T, Vitsupakorn, D, Saepua, S, Thongpanchang, C, Yuthavong, Y, Kamchonwongpaisan, S, Hoarau, M. | | Deposit date: | 2023-05-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of rigid biphenyl Plasmodium falciparum DHFR inhibitors using a fragment linking strategy.
Rsc Med Chem, 14, 2023
|
|
8JFD
 
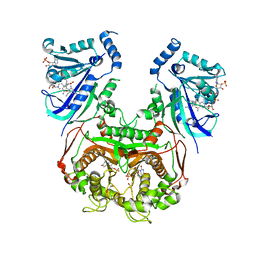 | | V1/S quadruple mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with compound 8 (B21594), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 3-[3-[[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]methyl]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Saeyang, T, Vitsupakorn, D, Saepua, S, Thongpanchang, C, Yuthavong, Y, Kamchonwongpaisan, S, Hoarau, M. | | Deposit date: | 2023-05-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of rigid biphenyl Plasmodium falciparum DHFR inhibitors using a fragment linking strategy.
Rsc Med Chem, 14, 2023
|
|
8RFN
 
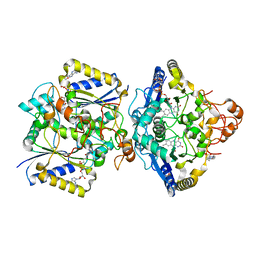 | | Human NOQ1 enzyme in its holo form by serial crystallography | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Martin-Garcia, J.M, Grieco, A, Medina, M, Boneta, S, Pey, A.L. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural dynamics and functional cooperativity of human NQO1 by ambient temperature serial crystallography and simulations.
Protein Sci., 33, 2024
|
|
5X7P
 
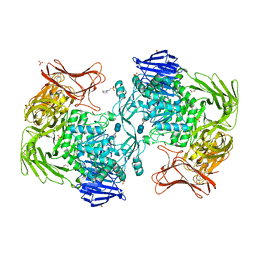 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
8RPJ
 
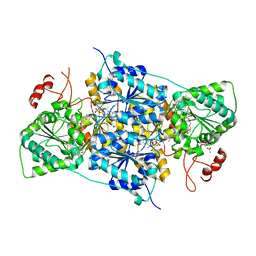 | | JanthE from Janthinobacterium sp. HH01 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RPI
 
 | | JanthE from Janthinobacterium sp. HH01, lactyl-ThDP | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8JIA
 
 | | Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsE(E165Q)X/RipC complex in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, Luo, M. | | Deposit date: | 2023-05-26 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Regulation of the cell division hydrolase RipC by the FtsEX system in Mycobacterium tuberculosis.
Nat Commun, 14, 2023
|
|
3DR0
 
 | | Structure of reduced cytochrome c6 from Synechococcus sp. PCC 7002 | | Descriptor: | Cytochrome c6, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Bialek, W, Krzywda, S, Jaskolski, M, Szczepaniak, A. | | Deposit date: | 2008-07-10 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Atomic-resolution structure of reduced cyanobacterial cytochrome c6 with an unusual sequence insertion
Febs J., 276, 2009
|
|
8IYB
 
 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
8J83
 
 | | Crystal structure of formate dehydrogenase from Methylorubrum extorquens AM1 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kobayashi, A, Taketa, M, Sowa, K, Kano, K, Higuchi, Y, Ogata, H. | | Deposit date: | 2023-04-29 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and function relationship of formate dehydrogenases: an overview of recent progress.
Iucrj, 10, 2023
|
|
7VXR
 
 | | Crystal structure of BPSL1038 from Burkholderia pseudomallei | | Descriptor: | BPSL1038, SODIUM ION | | Authors: | Shaibullah, S, Mohd-Sharif, M, Ho, K.L, Firdaus-Raih, M, Nathan, S, Mohamed, R, Teh, A.K, Waterman, J, Ng, C.L. | | Deposit date: | 2021-11-13 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and functional analyses of Burkholderia pseudomallei BPSL1038 reveal a Cas-2/VapD nuclease sub-family.
Commun Biol, 6, 2023
|
|
8IYC
 
 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
8IY8
 
 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Feruloyl esterase | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
7VXT
 
 | | Crystal structure of a selenomethionine-labeled BPSL1038 from Burkholderia pseudomallei | | Descriptor: | BETA-MERCAPTOETHANOL, BPSL1038, SODIUM ION | | Authors: | Shaibullah, S, Mohd-Sharif, M, Ho, K.L, Firdaus-Raih, M, Nathan, S, Mohamed, R, Teh, A.K, Waterman, J, Ng, C.L. | | Deposit date: | 2021-11-13 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and functional analyses of Burkholderia pseudomallei BPSL1038 reveal a Cas-2/VapD nuclease sub-family.
Commun Biol, 6, 2023
|
|
5Z9G
 
 | | Crystal structure of KAI2 | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
8J3S
 
 | | Complex structure of human cytomegalovirus protease and a macrocyclic peptide ligand | | Descriptor: | Assemblin, PHE-ILE-THR-GLY-HIS-TYR-TRP-VAL-ARG-PHE-LEU-PRO-CYS-GLY | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
8JE4
 
 | | Crystal structure of LimF prenyltransferase (H239G/W273T mutant) bound with the thiodiphosphate moiety of farnesyl S-thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, TRIHYDROGEN THIODIPHOSPHATE, prenyltransferase, ... | | Authors: | Hamada, K, Oguni, A, Zhang, Y, Satake, M, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Switching Prenyl Donor Specificities of Cyanobactin Prenyltransferases.
J.Am.Chem.Soc., 145, 2023
|
|
8IN4
 
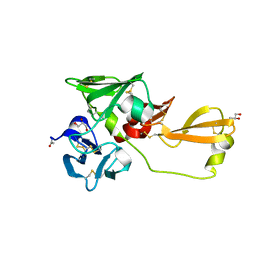 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, ACETYL GROUP, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8J3T
 
 | | Complex structure of human cytomegalovirus protease and a non-covalent small-molecule ligand | | Descriptor: | (4R)-1-[1-[(S)-[1-cyclopentyl-3-(2-methylphenyl)pyrazol-4-yl]-(4-methylphenyl)methyl]-2-oxidanylidene-pyridin-3-yl]-3-methyl-2-oxidanylidene-N-(3-oxidanylidene-2-azabicyclo[2.2.2]octan-4-yl)imidazolidine-4-carboxamide, Assemblin | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
8IN1
 
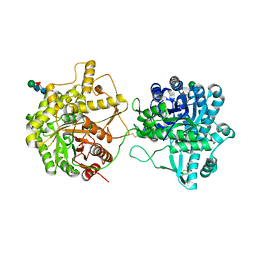 | | beta-glucosidase protein from Aplysia kurodai | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-Glucosidase, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8YA8
 
 | |
8IN3
 
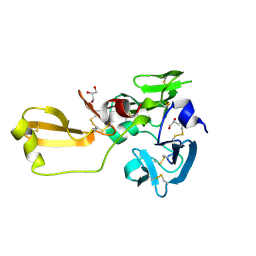 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
