7NPQ
 
 | |
4OYI
 
 | | Human solAC Complexed with (4-Amino-furazan-3-yl)-phenyl-methanone | | Descriptor: | (4-azanyl-1,2,5-oxadiazol-3-yl)-phenyl-methanone, Adenylate cyclase type 10, GLYCEROL | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-12 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
4OYW
 
 | | Crystal Structure of Human Soluble Adenylate Cyclase | | Descriptor: | Adenylate cyclase type 10, CHLORIDE ION, GLYCEROL | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
7NH6
 
 | | Crystal structure of human carbonic anhydrase II with 3-(3-((1-(2-(hydroxymethyl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-1H-1,2,3-triazol-4-yl)methyl)ureido)benzenesulfonamide | | Descriptor: | 3-(3-((1-(2-(hydroxymethyl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-1H-1,2,3-triazol-4-yl)methyl)ureido)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2021-02-10 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Mechanisms of the Antiproliferative and Antitumor Activity of Novel Telomerase-Carbonic Anhydrase Dual-Hybrid Inhibitors.
J.Med.Chem., 64, 2021
|
|
5X7R
 
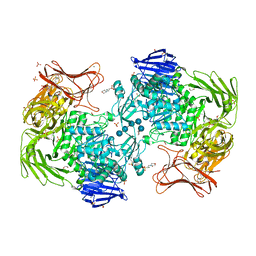 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with isomaltohexaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
7NH8
 
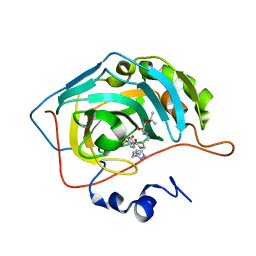 | | Crystal structure of human carbonic anhydrase II with N-((1-(6-((3aR,7R,7aS)-7-hydroxy-2,2-dimethyltetrahydro-[1,3]dioxolo[4,5-c]pyridin-5(4H)-yl)hexyl)-1H-1,2,3-triazol-4-yl)methyl)-4-sulfamoylbenzamide | | Descriptor: | Carbonic anhydrase 2, N-((1-(6-((3aR,7R,7aS)-7-hydroxy-2,2-dimethyltetrahydro-[1,3]dioxolo[4,5-c]pyridin-5(4H)-yl)hexyl)-1H-1,2,3-triazol-4-yl)methyl)-4-sulfamoylbenzamide, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2021-02-10 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Synthesis of Azasugar-Sulfonamide conjugates and their Evaluation as Inhibitors of Carbonic Anhydrases: the Azasugar Approach to Selectivity
Eur.J.Org.Chem., 2021
|
|
5XAS
 
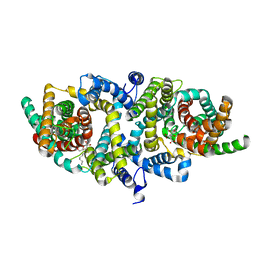 | | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS | | Descriptor: | CITRATE ANION, Citrate-sodium symporter, SODIUM ION, ... | | Authors: | Jin, M.S, Kim, J.W, Kim, S, Kim, S, Lee, H, Lee, J.-O. | | Deposit date: | 2017-03-14 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS
Sci Rep, 7, 2017
|
|
7N97
 
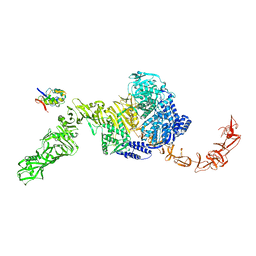 | | State 2 of TcdB and FZD2 at pH5 | | Descriptor: | Frizzled-2, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis for Receptor Recognition of the Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N8X
 
 | | Partial C. difficile TcdB and CSPG4 fragment | | Descriptor: | Chondroitin sulfate proteoglycan 4, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-16 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
5XGZ
 
 | | Metagenomic glucose-tolerant glycosidase | | Descriptor: | Beta-glycosidase, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Watanabe, M, Matsuzawa, T, Yaoi, K. | | Deposit date: | 2017-04-19 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Improved thermostability of a metagenomic glucose-tolerant beta-glycosidase based on its X-ray crystal structure.
Appl.Microbiol.Biotechnol., 101, 2017
|
|
7N9Q
 
 | | State 3 of TcdB and FZD2 at pH5 | | Descriptor: | Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7NFZ
 
 | |
7N9S
 
 | | TcdB and frizzled-2 CRD complex | | Descriptor: | Frizzled-2, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
4OIT
 
 | | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis | | Descriptor: | LysM domain protein, alpha-D-mannopyranose, beta-D-mannopyranose | | Authors: | Patra, D, Mishra, P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-01-20 | | Release date: | 2014-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis.
Glycobiology, 24, 2014
|
|
7N9R
 
 | | state 4 of TcdB and FZD2 at pH5 | | Descriptor: | Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
3AX8
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 15alpha-methoxy-1alpha,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3S,3aS,7aR)-1-[(2R)-6-hydroxy-6-methyl-heptan-2-yl]-3-methoxy-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-03-30 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New C15-substituted active vitamin D3
Org.Lett., 13, 2011
|
|
7N9Y
 
 | | Full-length TcdB and CSPG4 (401-560) complex | | Descriptor: | Chondroitin sulfate proteoglycan 4, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
3AXA
 
 | | Crystal structure of afadin PDZ domain in complex with the C-terminal peptide from nectin-3 | | Descriptor: | Afadin, Nectin-3 | | Authors: | Fujiwara, Y, Goda, N, Narita, H, Satomura, K, Nakagawa, A, Sakisaka, T, Suzuki, M, Hiroaki, H. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of afadin PDZ domain-nectin-3 complex shows the structural plasticity of the ligand-binding site.
Protein Sci., 24, 2015
|
|
4OOP
 
 | | Arabidopsis thaliana dUTPase with with magnesium and alpha,beta-imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Inoguchi, N, Bajaj, M, Moriyama, H. | | Deposit date: | 2014-02-03 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the mechanism defining substrate affinity in Arabidopsis thaliana dUTPase: the role of tryptophan 93 in ligand orientation.
BMC Res Notes, 8, 2015
|
|
7N95
 
 | | state 1 of TcdB and FZD2 at pH5 | | Descriptor: | Frizzled-2, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7NL8
 
 | |
7NHW
 
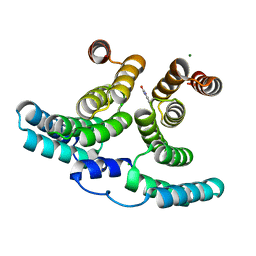 | |
8QAQ
 
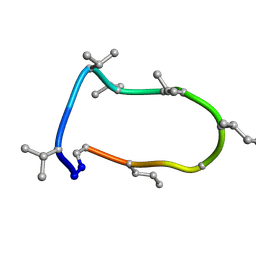 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. Conformation 1 of omphalotin A in apolar solvents. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8QAS
 
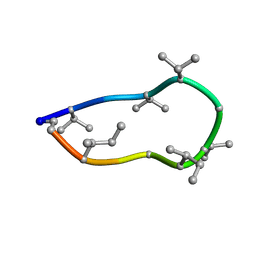 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
4O8F
 
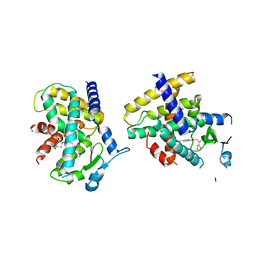 | | Crystal Structure of the complex between PPARgamma mutant R357A and rosiglitazone | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Chiaraluce, R, Consalvi, V, Lori, C, Loiodice, F, Laghezza, A, Pasquo, A, Cervoni, L, Aschi, M. | | Deposit date: | 2013-12-27 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the transactivation deficiency of the human PPAR gamma F360L mutant associated with familial partial lipodystrophy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
