123L
 
 | |
125L
 
 | |
126L
 
 | |
8SW5
 
 | |
128L
 
 | |
8SW6
 
 | |
127L
 
 | |
131D
 
 | | THE LOW-TEMPERATURE CRYSTAL STRUCTURE OF THE PURE-SPERMINE FORM OF Z-DNA REVEALS BINDING OF A SPERMINE MOLECULE IN THE MINOR GROOVE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SODIUM ION, SPERMINE | | Authors: | Bancroft, D, Williams, L.D, Rich, A, Egli, M. | | Deposit date: | 1993-06-18 | | Release date: | 1993-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The low-temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove.
Biochemistry, 33, 1994
|
|
1A80
 
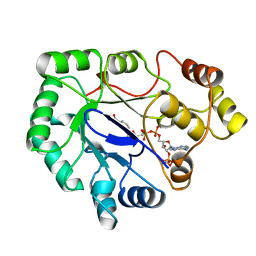 | | Native 2,5-DIKETO-D-GLUCONIC acid reductase a from CORYNBACTERIUM SP. complexed with nadph | | Descriptor: | 2,5-DIKETO-D-GLUCONIC ACID REDUCTASE A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Khurana, S, Powers, D.B, Anderson, S, Blaber, M. | | Deposit date: | 1998-03-31 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of 2,5-diketo-D-gluconic acid reductase A complexed with NADPH at 2.1-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1ACB
 
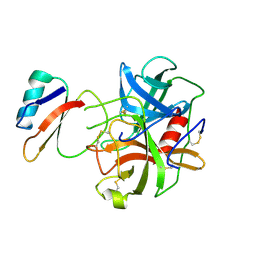 | | CRYSTAL AND MOLECULAR STRUCTURE OF THE BOVINE ALPHA-CHYMOTRYPSIN-EGLIN C COMPLEX AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | ALPHA-CHYMOTRYPSIN, Eglin C | | Authors: | Bolognesi, M, Frigerio, F, Coda, A, Pugliese, L, Lionetti, C, Menegatti, E, Amiconi, G, Schnebli, H.P, Ascenzi, P. | | Deposit date: | 1991-11-08 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal and molecular structure of the bovine alpha-chymotrypsin-eglin c complex at 2.0 A resolution.
J.Mol.Biol., 225, 1992
|
|
138D
 
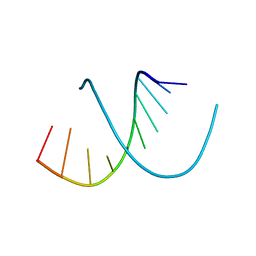 | | A-DNA DECAMER D(GCGGGCCCGC)-HEXAGONAL CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
137D
 
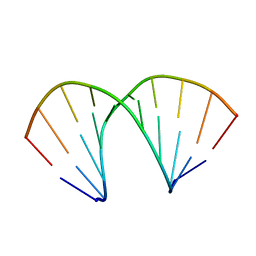 | | A-DNA DECAMER D(GCGGGCCCGC)-ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
137L
 
 | |
6FV4
 
 | | The structure of N-acetyl-D-glucosamine-6-phosphate deacetylase D267A mutant from Mycobacterium smegmatis in complex with N-acetyl-D-glucosamine-6-phosphate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, CADMIUM ION, ... | | Authors: | Ahangar, M.S, Furze, C.M, Guy, C.S, Cooper, C, Maskew, K.S, Graham, B, Cameron, A.D, Fullam, E. | | Deposit date: | 2018-03-01 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structural and functional determination of homologs of theMycobacterium tuberculosis N-acetylglucosamine-6-phosphate deacetylase (NagA).
J. Biol. Chem., 293, 2018
|
|
6FZM
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor ITK6 | | Descriptor: | 4-[(8-methyl-4-oxidanylidene-7-prop-1-ynyl-3~{H}-quinazolin-2-yl)methylsulfanyl]benzoic acid, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Kirby, I.T, Sreenivasan, R, Cohen, M.S, Schuler, H. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | A Potent and Selective PARP11 Inhibitor Suggests Coupling between Cellular Localization and Catalytic Activity.
Cell Chem Biol, 25, 2018
|
|
6Z93
 
 | | [4Fe-4S]-dependent thiouracil desulfidase TudS (DUF523Vcz) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Pecqueur, L, Zhou, J, Fontecave, M, Golinelli-Pimpaneau, B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Structural Evidence for a [4Fe-5S] Intermediate in the Non-Redox Desulfuration of Thiouracil.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6Z92
 
 | | [4Fe-4S]-dependent thiouracil desulfidase TudS (DUF523Vcz) solved by Fe-SAD phasing | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DUF523 domain-containing protein, ... | | Authors: | Pecqueur, L, Zhou, J, Fontecave, M, Golinelli-Pimpaneau, B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | Structural Evidence for a [4Fe-5S] Intermediate in the Non-Redox Desulfuration of Thiouracil.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6GCI
 
 | | Structure of the bongkrekic acid-inhibited mitochondrial ADP/ATP carrier | | Descriptor: | Bongkrekic acid, CARDIOLIPIN, HEXAETHYLENE GLYCOL, ... | | Authors: | Ruprecht, J.J, King, M.S, Pardon, E, Aleksandrova, A.A, Zogg, T, Crichton, P.G, Steyaert, J, Kunji, E.R.S. | | Deposit date: | 2018-04-17 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Molecular Mechanism of Transport by the Mitochondrial ADP/ATP Carrier.
Cell, 176, 2019
|
|
6GDY
 
 | | Crystal structure of 2OG oxygenase JMJD6 (aa 1-343) in complex with Fe(II) and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Islam, M.S, Schofield, C.J, McDonough, M.A. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Biochemical and structural investigations clarify the substrate selectivity of the 2-oxoglutarate oxygenase JMJD6.
J.Biol.Chem., 294, 2019
|
|
6ZJB
 
 | | Crystal structure of human adenylate kinase 3, AK3, in complex with inhibitor Gp5A | | Descriptor: | GTP:AMP phosphotransferase AK3, mitochondrial, MAGNESIUM ION, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Structural Basis for GTP versus ATP Selectivity in the NMP Kinase AK3.
Biochemistry, 59, 2020
|
|
6FU2
 
 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Psychrobacter arcticus in complex with PRPP and ATP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2018-02-26 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6FU7
 
 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Psychrobacter arcticus in complex with PRATP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, ... | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2018-02-26 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6ZJD
 
 | | Crystal structure of human adenylate kinase 3, AK3, in complex with inhibitor ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GTP:AMP phosphotransferase AK3, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for GTP versus ATP Selectivity in the NMP Kinase AK3.
Biochemistry, 59, 2020
|
|
6Z94
 
 | | [4Fe-4S]-dependent thiouracil desulfidase TudS (DUF523Vcz)(S-SAD data) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DUF523 domain-containing protein, ... | | Authors: | Pecqueur, L, Zhou, J, Fontecave, M, Golinelli-Pimpaneau, B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Structural Evidence for a [4Fe-5S] Intermediate in the Non-Redox Desulfuration of Thiouracil.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6ZJE
 
 | | Crystal structure of human adenylate kinase 3, AK3, in complex with inhibitor Ap5A | | Descriptor: | BIS(ADENOSINE)-5'-PENTAPHOSPHATE, CHLORIDE ION, GTP:AMP phosphotransferase AK3, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Basis for GTP versus ATP Selectivity in the NMP Kinase AK3.
Biochemistry, 59, 2020
|
|
