7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
2AWH
 
 | | Human Nuclear Receptor-Ligand Complex 1 | | Descriptor: | Peroxisome proliferator activated receptor delta, VACCENIC ACID, heptyl beta-D-glucopyranoside | | Authors: | Fyffe, S.A, Alphey, M.S, Buetow, L, Smith, T.K, Ferguson, M.A.J, Sorensen, M.D, Bjorkling, F, Hunter, W.N. | | Deposit date: | 2005-09-01 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant Human PPAR-beta/delta Ligand-binding Domain is Locked in an Activated Conformation by Endogenous Fatty Acids
J.Mol.Biol., 356, 2006
|
|
2CHO
 
 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity | | Descriptor: | ACETATE ION, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
2BAW
 
 | | Human Nuclear Receptor-Ligand Complex 1 | | Descriptor: | Peroxisome proliferator activated receptor delta, VACCENIC ACID, heptyl beta-D-glucopyranoside | | Authors: | Fyffe, S.A, Alphey, M.S, Buetow, L, Smith, T.K, Ferguson, M.A.J, Sorensen, M.D, Bjorkling, F, Hunter, W.N. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reevaluation of the PPAR-beta/delta Ligand Binding Domain Model Reveals Why It Exhibits the Activated Form
Mol.Cell, 21, 2006
|
|
1ZWT
 
 | | Structure of the globular head domain of the bundlin, BfpA, of the bundle-forming pilus of Enteropathogenic E.coli | | Descriptor: | Major structural subunit of bundle-forming pilus | | Authors: | Ramboarina, S, Fernandes, P.J, Daniell, S, Islam, S, Frankel, G, Booy, F, Donnenberg, M.S, Matthews, S. | | Deposit date: | 2005-06-06 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the Bundle-forming Pilus from Enteropathogenic Escherichia coli
J.Biol.Chem., 280, 2005
|
|
2A7B
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | XENON, gamma-adaptin appendage domain | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A7I
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A7H
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cationic trypsin | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2CIO
 
 | |
2C7Z
 
 | | Plant enzyme crystal form II | | Descriptor: | 3-KETOACYL-COA THIOLASE 2 | | Authors: | Sundaramoorthy, R, Micossi, E, Alphey, M.S, Leonard, G.A, Hunter, W.N. | | Deposit date: | 2005-11-30 | | Release date: | 2006-05-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Crystal Structure of a Plant 3-Ketoacyl-Coa Thiolase Reveals the Potential for Redox Control of Peroxisomal Fatty Acid Beta-Oxidation.
J.Mol.Biol., 359, 2006
|
|
2C7Y
 
 | | plant enzyme | | Descriptor: | 3-KETOACYL-COA THIOLASE 2 | | Authors: | Sundaramoorthy, R, Micossi, E, Alphey, M.S, Germain, V, Bryce, J.H, Smith, S.M, Leonard, G.A, Hunter, W.N. | | Deposit date: | 2005-11-30 | | Release date: | 2006-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a Plant 3-Ketoacyl-Coa Thiolase Reveals the Potential for Redox Control of Peroxisomal Fatty Acid Beta-Oxidation.
J.Mol.Biol., 359, 2006
|
|
9AXO
 
 | |
6QGA
 
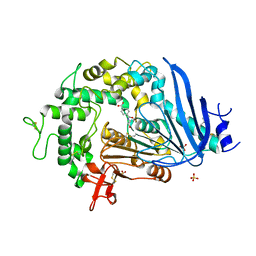 | | Crystal structure of Ideonella sakaiensis MHETase bound to the non-hydrolyzable ligand MHETA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-hydroxyethylcarbamoyl)benzoic acid, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
7OEX
 
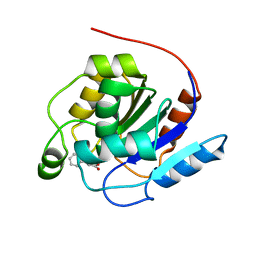 | |
9FLU
 
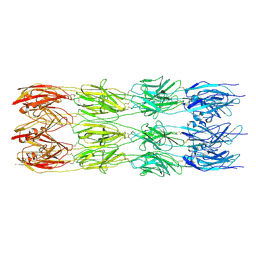 | | CryoEM structure of the fragment-1 (3402-3733) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
7NTN
 
 | | The structure of RRM domain of human TRMT2A at 2 A resolution | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Davydova, E, Janowski, R, Witzenberger, M, Niessing, D. | | Deposit date: | 2021-03-10 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Small-molecule modulators of TRMT2A decrease PolyQ aggregation and PolyQ-induced cell death.
Comput Struct Biotechnol J, 20, 2022
|
|
7OFX
 
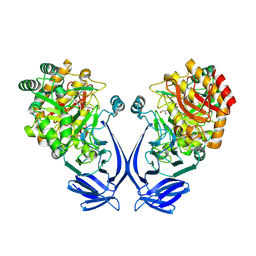 | | Crystal structure of a GH31 family sulfoquinovosidase mutant D455N from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | Alpha-glucosidase yihQ, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7OFY
 
 | | Crystal structure of SQ binding protein from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | 1,2-ETHANEDIOL, Sulfoquinovosyl binding protein, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Jarva, M.A, Sharma, M, Goddard-Borger, E.D, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9EP9
 
 | | NMR solution structure of lipid transfer protei Sola l7 from tomato seeds | | Descriptor: | Non-specific lipid-transfer protein | | Authors: | Parron-Ballesteros, J, Mantin-Pedraz, L, G.Gordo, R, Mayorga, C, Villaba, M, Batanero, E, Pantoja-Uceda, D, Turnay, J. | | Deposit date: | 2024-03-18 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Long-chain fatty acids block allergic reaction against lipid transfer protein Sola l 7 from tomato seeds.
Protein Sci., 33, 2024
|
|
9AXQ
 
 | |
7NTO
 
 | | The structure of RRM domain of human TRMT2A at 1.23 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Witzenberger, M, Janowski, R, Davydova, E, Niessing, D. | | Deposit date: | 2021-03-10 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Small-molecule modulators of TRMT2A decrease PolyQ aggregation and PolyQ-induced cell death.
Comput Struct Biotechnol J, 20, 2022
|
|
9AXN
 
 | |
9FSV
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with epoxyketone inhibitor 42 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-22 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
4Q3N
 
 | | Crystal structure of MGS-M5, a lactate dehydrogenase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
9AXR
 
 | |
