7P8E
 
 | | Crystal structure of the Receiver domain of M. truncatula cytokinin receptor MtCRE1 | | Descriptor: | CALCIUM ION, Receiver domain of histidine kinase | | Authors: | Tran, L.H, Urbanowicz, A, Jasinski, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3D Domain Swapping Dimerization of the Receiver Domain of Cytokinin Receptor CRE1 From Arabidopsis thaliana and Medicago truncatula .
Front Plant Sci, 12, 2021
|
|
9AXQ
 
 | |
9AXP
 
 | |
9FSV
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with epoxyketone inhibitor 42 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-22 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9FLX
 
 | | CryoEM structure of the fragment-4 (4074-4421) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9G8B
 
 | | CryoEM structure of the fragment-2 (2892-3236) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9AXR
 
 | |
9FLV
 
 | | CryoEM structure of the fragment-5 (2393-2807) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9FLS
 
 | | CryoEM structure of the fragment-3 (2061-2397) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9FLW
 
 | | CryoEM structure of the fragment-6 (3571-4078) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9AXO
 
 | |
9FLP
 
 | | CryoEM structure of the neck (1403-2314) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9AXN
 
 | |
9FLY
 
 | | CryoEM structure of the base (4151-5009) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
5ONJ
 
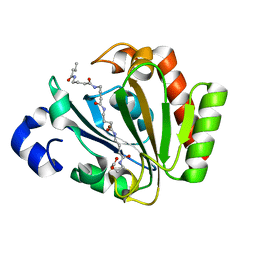 | | YnDL in Complex with 5 amino acid (PGA) complex | | Descriptor: | (2R,3S,4R,5R,6R)-6-((1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYLOXY)-5-AMINO-2-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, YndL, ZINC ION | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
Febs J., 285, 2018
|
|
1HYP
 
 | |
7OT5
 
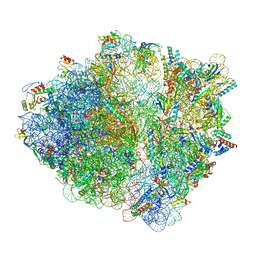 | | CspA-70 cotranslational folding intermediate 1 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-06-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OIF
 
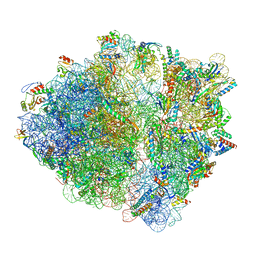 | | CspA-27 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
5OW6
 
 | | CryoEM structure of recombinant CMV particles with Tetanus-epitope | | Descriptor: | Capsid protein, VP2, VP3, ... | | Authors: | Kotecha, A, Stuart, D.I, Backmann, M. | | Deposit date: | 2017-08-30 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Incorporation of tetanus-epitope into virus-like particles achieves vaccine responses even in older recipients in models of psoriasis, Alzheimer's and cat allergy.
NPJ Vaccines, 2, 2017
|
|
8Q8N
 
 | | Crystal structure of human GPX4-U46C-I129S-L130S | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Bostock, M.J, Mourao, A, Napolitano, V, Sattler, M, Conrad, M, Popowicz, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An ultra-rare variant of GPX4 reveals the structural basis to avert neurodegeneration
To Be Published
|
|
1HYB
 
 | | CRYSTAL STRUCTURE OF AN ACTIVE SITE MUTANT OF METHANOBACTERIUM THERMOAUTOTROPHICUM NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Saridakis, V, Christendat, D, Kimber, M.S, Edwards, A.M, Pai, E.F. | | Deposit date: | 2001-01-18 | | Release date: | 2001-03-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.
J.Biol.Chem., 276, 2001
|
|
7OMT
 
 | | Crystal structure of ProMacrobody 21 with bound maltose | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, ProMacrobody 21, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
1HVV
 
 | |
7MJB
 
 | | Crystal Structure of Nanoluc Luciferase Mutant R164Q | | Descriptor: | CHLORIDE ION, DECANOIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Reza, M.S, Ai, H, Minor, W. | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Nanoluc Luciferase Mutant R164Q
To Be Published
|
|
4RIL
 
 | | Structure of the amyloid forming segment, GAVVTGVTAVA, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 68-78, determined by electron diffraction | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.43 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
