6B64
 
 | | IMPase (AF2372) with 25 mM Asp | | Descriptor: | ASPARTIC ACID, Fructose-1,6-bisphosphatase/inositol-1-monophosphatase, MAGNESIUM ION, ... | | Authors: | Goldstein, R.I, Roberts, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Osmolyte binding capacity of a dual action IMPase/FBPase (AF2372)
To Be Published
|
|
8B7J
 
 | | Human HSP90 alpha ATP Binding Domain, ATP-lid closed conformation, R46A | | Descriptor: | HSP90AA1 protein | | Authors: | Rioual, E, Henot, F, Favier, A, Macek, P, Crublet, E, Josso, P, Brustcher, B, Frech, M, Gans, P, Loison, C, Boisbouvier, J. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Visualizing the transiently populated closed-state of human HSP90 ATP binding domain.
Nat Commun, 13, 2022
|
|
4A5B
 
 | |
4A7Z
 
 | | Complex of bifunctional aldos-2-ulose dehydratase with the reaction intermediate ascopyrone M | | Descriptor: | ALDOS-2-ULOSE DEHYDRATASE, Ascopyrone M, MAGNESIUM ION, ... | | Authors: | Claesson, M, Lindqvist, Y, Madrid, S, Sandalova, T, Fiskesund, R, Yu, S, Schneider, G. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Bifunctional Aldos-2-Ulose Dehydratase/Isomerase from Phanerochaete Chrysosporium with the Reaction Intermediate Ascopyrone M.
J.Mol.Biol., 417, 2012
|
|
6VGW
 
 | | Crystal structure of VidaL intein (selenomethionine variant) | | Descriptor: | GLYCEROL, SULFATE ION, VidaL | | Authors: | Burton, A.J, Haugbro, M, Parisi, E, Muir, T.W. | | Deposit date: | 2020-01-09 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Live-cell protein engineering with an ultra-short split intein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5G4G
 
 | | Structure of the ATPgS-bound VAT complex | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4AAX
 
 | | CpGH89CBM32-5, from Clostridium perfringens, in complex with N- acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-12-05 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
6VAR
 
 | | 61 nt human Hepatitis B virus epsilon pre-genomic RNA | | Descriptor: | RNA (61-MER) | | Authors: | LeBlanc, R.M, Kasprzak, W.K, Longhini, A.P, Abulwerdi, F, Ginocchio, S, Shields, B, Nyman, J, Svirydava, M, Del Vecchio, C, Ivanic, J, Schneekloth, J.S, Dayie, T.K, Shapiro, B.A, Le Grice, S.F.J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structural insights of the conserved "priming loop" of hepatitis B virus pre-genomic RNA.
J.Biomol.Struct.Dyn., 2021
|
|
5G64
 
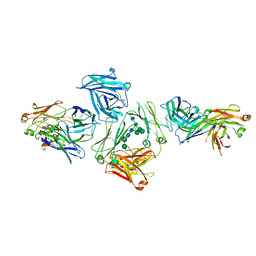 | | The complex between human IgE-Fc and two anti-IgE Fab fragments | | Descriptor: | FAB FRAGMENT, IG EPSILON CHAIN C REGION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Davies, A.M, Allan, E.G, Keeble, A.H, Delgado, J, Cossins, B.P, Mitropoulou, A.N, Pang, M.O.Y, Ceska, T, Beavil, A.J, Craggs, G, Westwood, M, Henry, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-06-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.715 Å) | | Cite: | Allosteric mechanism of action of the therapeutic anti-IgE antibody omalizumab.
J. Biol. Chem., 292, 2017
|
|
5NOS
 
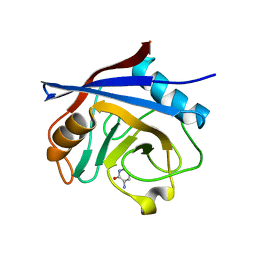 | | Structure of cyclophilin A in complex with 3-amino-1H-pyridin-2-one | | Descriptor: | 5-azanyl-3~{H}-pyridin-6-one, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
3NJY
 
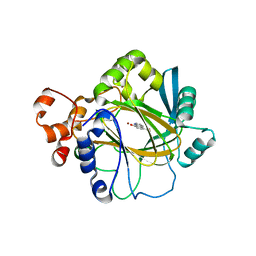 | | Crystal structure of JMJD2A complexed with 5-carboxy-8-hydroxyquinoline | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | King, O.N.F, Clifton, I.J, Wang, M, Maloney, D.J, Jadhav, A, Oppermann, U, Heightman, T.D, Simeonov, A, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2010-06-18 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors
Plos One, 5, 2010
|
|
5NOQ
 
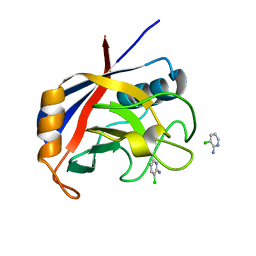 | | Structure of cyclophilin A in complex with 3-chloropyridin-2-amine | | Descriptor: | 3-chloranylpyridin-2-amine, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
5NOU
 
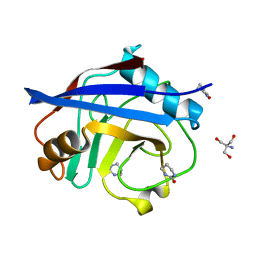 | | Structure of cyclophilin A in complex with hexahydropyrimidin-2-one | | Descriptor: | 1,3-diazinan-2-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
1VEA
 
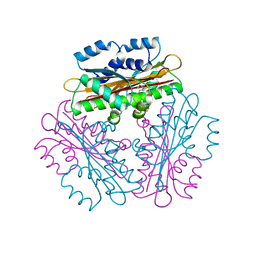 | | Crystal Structure of HutP, an RNA binding antitermination protein | | Descriptor: | Hut operon positive regulatory protein, N-(2-NAPHTHYL)HISTIDINAMIDE | | Authors: | Kumarevel, T.S, Fujimoto, Z, Karthe, P, Oda, M, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Activated HutP; An RNA Binding Protein that Regulates Transcription of the hut Operon in Bacillus subtilis
Structure, 12, 2004
|
|
5GHL
 
 | | Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger | | Descriptor: | GLYCEROL, Glucoamylase, SULFATE ION | | Authors: | Miyake, H, Suyama, Y, Muraki, N, Kusunoki, M, Tanaka, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the starch-binding domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
6V8M
 
 | | Crystal structure of Ara h 8.0201 | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, Ara h 8 allergen isoform, SULFATE ION | | Authors: | Pote, S, Offermann, L.R, Hurlburt, B.K, McBride, J.K, Chruszcz, M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Ara h 8.0201
To Be Published
|
|
5GX3
 
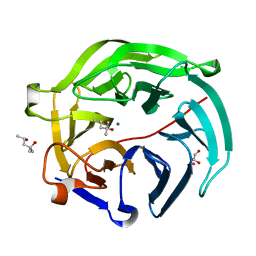 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 6.9 MGy (6th measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
4ACU
 
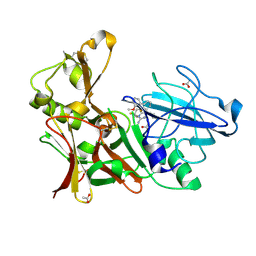 | | Aminoimidazoles as BACE-1 Inhibitors. X-RAY CRYSTAL STRUCTURE OF BETA SECRETASE COMPLEXED WITH COMPOUND 14 | | Descriptor: | (8S)-3,3-DIFLUORO-8-(2'-FLUORO-3'-METHOXYBIPHENYL-3-YL)-8-PYRIDIN-4-YL-2,3,4,8-TETRAHYDROIMIDAZO[1,5-A]PYRIMIDIN-6-AMINE, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B, Holenz, J, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Plobeck, N, Rotticci, D, Sehgelmeble, F, Sundstrom, M, von Berg, S, Falting, J, Georgievska, B, Gustavsson, S, Neelissen, J, Ek, M, Olsson, L.L, Berg, S. | | Deposit date: | 2011-12-19 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aminoimidazoles as BACE-1 inhibitors: the challenge to achieve in vivo brain efficacy.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
4AQD
 
 | | Crystal structure of fully glycosylated human butyrylcholinesterase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Wandhammer, M, Ronco, C, Trovaslet, M, Jean, L, Lockridge, O, Renard, P.Y, Nachon, F. | | Deposit date: | 2012-04-16 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human butyrylcholinesterase produced in insect cells: huprine-based affinity purification and crystal structure.
FEBS J., 279, 2012
|
|
8B10
 
 | | Crystal Structure of Shank2-SAM mutant domain - L1800W | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bento, I, Gracia Alai, M, Kreienkamp, J.-H. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-30 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural deficits in key domains of Shank2 lead to alterations in postsynaptic nanoclusters and to a neurodevelopmental disorder in humans.
Mol Psychiatry, 29, 2024
|
|
5AJO
 
 | | Crystal structure of the inactive form of GalNAc-T2 in complex with the glycopeptide MUC5AC-3,13 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUCIN, POLYPEPTIDE N-ACETYLGALACTOSAMINYLTRANSFERASE 2, ... | | Authors: | Lira-Navarrete, E, delasRivas, M, Companon, I, Pallares, M.C, Kong, Y, Iglesias-Fernandez, J, Bernardes, G.J.L, Peregrina, J.M, Rovira, C, Bernado, P, Bruscolini, P, Clausen, H, Lostao, A, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dynamic Interplay between Catalytic and Lectin Domains of Galnac-Transferases Modulates Protein O-Glycosylation.
Nat.Commun., 6, 2015
|
|
8B3T
 
 | | Hen Egg White Lysozyme 4s in situ crystallization | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
8B3U
 
 | | Hen Egg White Lysozyme 6s in situ crystallization | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
4AFZ
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, D(-)-TARTARIC ACID, FYNOMER | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
8B3V
 
 | | Hen Egg White Lysozyme 8s in situ crystallization | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
