2MV9
 
 | | Solution structure of Ovis Aries PrP with mutation delta193-196 | | Descriptor: | Major prion protein | | Authors: | Munoz, C, Egalon, A, Beringue, V, Rezaei, H, Dron, M, Sizun, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Generating Bona Fide Mammalian Prions with Internal Deletions.
J.Virol., 90, 2016
|
|
6C2W
 
 | | Crystal structure of human prothrombin mutant S101C/A470C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Prothrombin, ... | | Authors: | Chinnaraj, M, Chen, Z, Pelc, L, Grese, Z, Bystranowska, D, Di Cera, E, Pozzi, N. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.12 Å) | | Cite: | Structure of prothrombin in the closed form reveals new details on the mechanism of activation.
Sci Rep, 8, 2018
|
|
6BZU
 
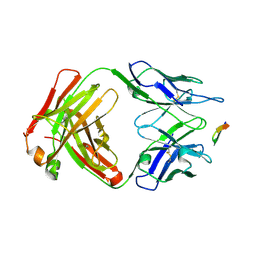 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody 19B3 | | Descriptor: | 19B3 Heavy Chain, 19B3 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BZY
 
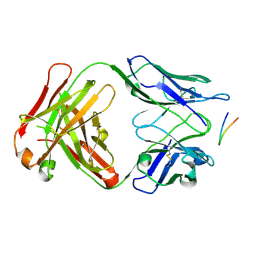 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the 22D11 broadly neutralizing antibody | | Descriptor: | 22D11 Heavy Chain, 22D11 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5UA3
 
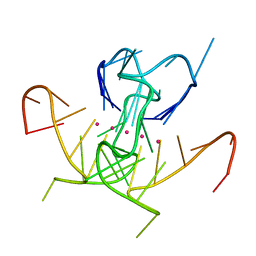 | | Crystal Structure of a DNA G-Quadruplex with a Cytosine Bulge | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*GP*CP*GP*GP*AP*GP*GP*GP*TP*GP*GP*GP*CP*CP*T)-3'), POTASSIUM ION | | Authors: | Meier, M, McDougall, M.D, Krahn, N.J, Moya-Torres, A, McRae, E.K.S, Booy, E.P, Patel, T.R, McKenna, S.A, Stetefeld, J. | | Deposit date: | 2016-12-18 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8802 Å) | | Cite: | Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge.
Nucleic Acids Res., 46, 2018
|
|
6C1V
 
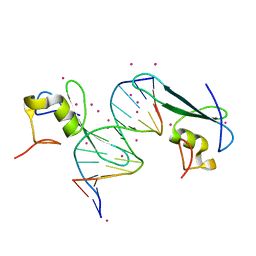 | | MBD2 in complex with double-stranded DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA.
J. Biol. Chem., 293, 2018
|
|
2N8D
 
 | |
6C7Z
 
 | | Crystal structure of the Q108K:K40L:T51V:R58F mutant of human Cellular Retinol Binding Protein II in complex with synthetic Ligand Julolidine | | Descriptor: | (2E,4E)-3-methyl-5-(2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-9-yl)penta-2,4-dienal, ACETATE ION, Retinol-binding protein 2 | | Authors: | Nosrati, M, Geiger, J.H. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A Genetically Encoded Ratiometric pH Probe: Wavelength Regulation-Inspired Design of pH Indicators.
Chembiochem, 19, 2018
|
|
4MML
 
 | | D40A Hfq from Pseudomonas aeruginosa | | Descriptor: | MAGNESIUM ION, Protein hfq, SULFATE ION, ... | | Authors: | Murina, V.N, Filimonov, V.V, Melnik, B.S, Uhlein, M, Mueller, U, Weiss, M, Nikulin, A.D. | | Deposit date: | 2013-09-09 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Effect of conserved intersubunit amino Acid substitutions on hfq protein structure and stability.
Biochemistry Mosc., 79, 2014
|
|
6BOM
 
 | |
6BQH
 
 | | Crystal structure of 5-HT2C in complex with ritanserin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, 6-(2-{4-[bis(4-fluorophenyl)methylidene]piperidin-1-yl}ethyl)-7-methyl-5H-[1,3]thiazolo[3,2-a]pyrimidin-5-one, ... | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
6FIV
 
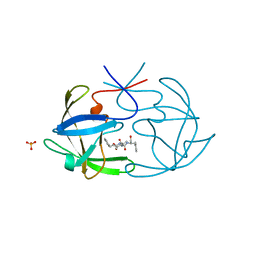 | | STRUCTURAL STUDIES OF HIV AND FIV PROTEASES COMPLEXED WITH AN EFFICIENT INHIBITOR OF FIV PR | | Descriptor: | RETROPEPSIN, SULFATE ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Li, M, Lee, T, Morris, G, Laco, G, Wong, C, Olson, A, Elder, J, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-12-02 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease
Proteins, 38, 2000
|
|
5OJN
 
 | |
6BV8
 
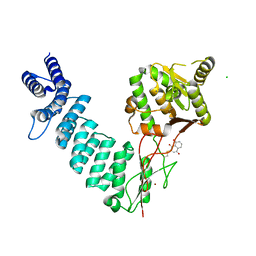 | | Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana complexed with Mn after 3-hour soak with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Karasik, A, Wu, N, Fierke, C.A, Koutmos, M. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of protein-only RNase P with Gambogic acid and Juglone
To Be Published
|
|
2MTX
 
 | |
6HLK
 
 | | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. | | Descriptor: | Redirecting phage packaging protein C (RppC) | | Authors: | Penades, J.R, Bacarizo, J, Marina, A, Alqasmi, M, Fillol-Salom, A, Roszak, A.W, Ciges-Tomas, J.R. | | Deposit date: | 2018-09-11 | | Release date: | 2019-07-31 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit.
Mol.Cell, 75, 2019
|
|
2MQH
 
 | |
6C8H
 
 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of gadolinium | | Descriptor: | GADOLINIUM ATOM, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5U5X
 
 | | CcP gateless cavity | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Peroxidase, THIOPHENE-3-CARBOXIMIDAMIDE | | Authors: | Fischer, M, Shoichet, B.K. | | Deposit date: | 2016-12-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Testing inhomogeneous solvation theory in structure-based ligand discovery.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3M58
 
 | | SET7/9 Y245A in complex with TAF10-K189me1 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
2MWG
 
 | |
3M5A
 
 | | SET7/9 Y245A in complex with TAF10-K189me3 peptide and AdoHcy | | Descriptor: | COBALT (II) ION, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
5OGN
 
 | | Metalacarborane inhibitors of Carbonic Anhydrase IX | | Descriptor: | Carbonic anhydrase 2, ZINC ION, cobaltcarborane | | Authors: | Brynda, J, Rezacova, P, Pospisilova, K, Kugler, M, Gruner, B, Sicha, V. | | Deposit date: | 2017-07-13 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Metallacarborane Sulfamides: Unconventional, Specific, and Highly Selective Inhibitors of Carbonic Anhydrase IX.
J.Med.Chem., 2019
|
|
6FJY
 
 | | Crystal structure of CsuC-CsuE chaperone-tip adhesion subunit pre-assembly complex from archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuC, Protein CsuE | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-16 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis forAcinetobacter baumanniibiofilm formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3M7Q
 
 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean sea anemone stichodactyla helianthus in complex with bovine pancreatic trypsin | | Descriptor: | Cationic trypsin, Kunitz-type proteinase inhibitor SHPI-1, PHOSPHATE ION | | Authors: | Garcia-Fernandez, R, Redecke, L, Pons, T, Perbandt, M, Gil, D, Talavera, A, Gonzalez, Y, de los angeles Chavez, M, Betzel, C. | | Deposit date: | 2010-03-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into serine protease inhibition by a marine invertebrate BPTI Kunitz-type inhibitor.
J.Struct.Biol., 180, 2012
|
|
