6B07
 
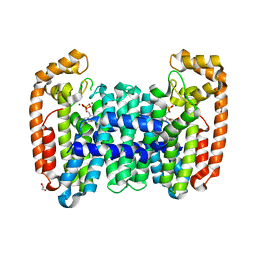 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with [1-phosphono-2-(1-propylpyridin-2-yl)ethyl]phosphonic acid (inhibitor 1d) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,2-diphosphonoethyl)-1-propylpyridin-1-ium, Farnesyl diphosphate synthase, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
5TVH
 
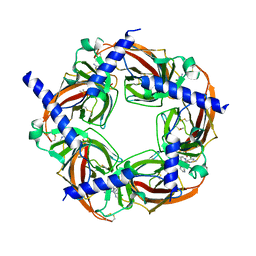 | | Crystal structure of AChBP from Aplysia californica complex with 2-aminopyrimidine at pH 8.0 spacegroup P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-N~4~,N~4~-bis[(pyridin-3-yl)methyl]pyrimidine-2,4-diamine, DIMETHYL SULFOXIDE, ... | | Authors: | Camacho-Hernandez, G.A, Kaczanowska, K, Harel, M, Finn, M.G, Taylor, P.W. | | Deposit date: | 2016-11-08 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of AChBP from Aplysia californica complex with 2-aminopyrimidine at pH 7.0 spacegroup P212121
To Be Published
|
|
6EB2
 
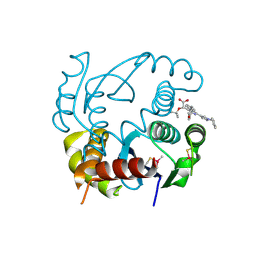 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-[1-(1-benzyl-1H-pyrazol-4-yl)-3-(3,4-dihydro-2H-1-benzopyran-6-yl)isoquinolin-4-yl](tert-butoxy)acetic acid | | Descriptor: | (2S)-[1-(1-benzyl-1H-pyrazol-4-yl)-3-(3,4-dihydro-2H-1-benzopyran-6-yl)isoquinolin-4-yl](tert-butoxy)acetic acid, Integrase | | Authors: | Lindenberger, J.J, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | An Isoquinoline Scaffold as a Novel Class of Allosteric HIV-1 Integrase Inhibitors.
ACS Med Chem Lett, 10, 2019
|
|
6J7W
 
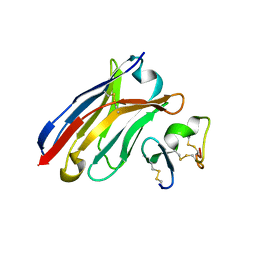 | | Crystal Structure of Human BCMA in complex with UniAb(TM) VH | | Descriptor: | Tumor necrosis factor receptor superfamily member 17, UniAb | | Authors: | Clarke, S.C, Ma, B, Trinklein, N.D, Schellenberger, U, Osborn, M, Ouisse, L, Boudreau, A, Davison, L, Harris, K.E, Ugamraj, H, Balasubramani, A, Dang, K, Jorgensen, B, Ogana, H, Pham, D, Pratap, P, Sankaran, P, Anegon, I, van Schooten, W, Bruggemann, M, Buelow, R, Force Aldred, S. | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Multispecific Antibody Development Platform Based on Human Heavy Chain Antibodies
Front Immunol, 9, 2018
|
|
6PEV
 
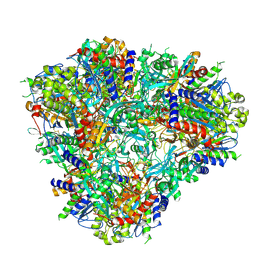 | |
7MLH
 
 | | Crystal structure of human IgE (2F10) in complex with Der p 2.0103 | | Descriptor: | CHLORIDE ION, Der p 2 variant 3, IgE Heavy chain, ... | | Authors: | Khatri, K, Kapingidza, A.B, Richardson, C.M, Vailes, L.D, Wunschmann, S, Dolamore, C, Chapman, M.D, Smith, S.A, Pomes, A, Chruszcz, M. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human IgE monoclonal antibody recognition of mite allergen Der p 2 defines structural basis of an epitope for IgE cross-linking and anaphylaxis in vivo.
Pnas Nexus, 1, 2022
|
|
6E1L
 
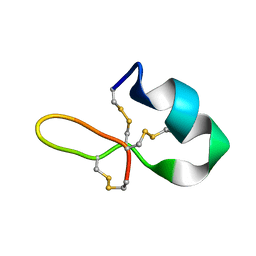 | | GRN3Ala | | Descriptor: | Granulin | | Authors: | Dastpeyman, M, Bansal, P, Wilson, D, Sotillo, J, Brindley, P, Loukas, A, Smout, M, Daly, N. | | Deposit date: | 2018-07-10 | | Release date: | 2019-02-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Variants of a Liver Fluke Derived Granulin Peptide Potently Stimulate Wound Healing.
J. Med. Chem., 61, 2018
|
|
6B06
 
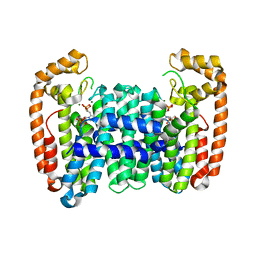 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with IPP and [2-(1-methylpyridin-2-yl)-1-phosphono-ethyl]phosphonic acid (inhibitor 1b) | | Descriptor: | 2-(2,2-diphosphonoethyl)-1-methylpyridin-1-ium, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl diphosphate synthase, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
5U20
 
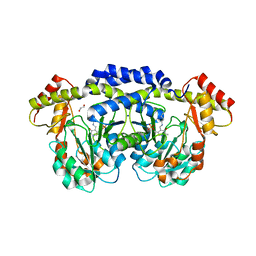 | | X-ray structure of the WlaRG aminotransferase from Campylobacter jejuni, internal PLP-aldimine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative aminotransferase, ... | | Authors: | Thoden, J.B, Holden, H.M, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
6PIM
 
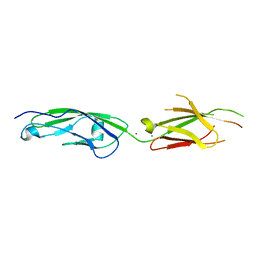 | | Crystal Structure of Human Protocadherin-1 EC3-4 | | Descriptor: | CALCIUM ION, Protocadherin-1 | | Authors: | Modak, D, Sotomayor, M. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Identification of an adhesive interface for the non-clustered delta 1 protocadherin-1 involved in respiratory diseases.
Commun Biol, 2, 2019
|
|
6YLR
 
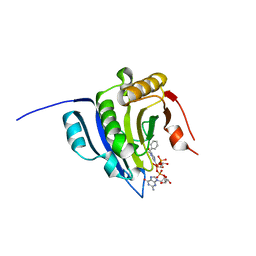 | | Translation initiation factor 4E in complex with bn7GpppG mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, bn7GpppG mRNA 5' cap analog | | Authors: | Kubacka, D, Wojcik, R, Baranowski, M.R, Warminski, M, Kowalska, J, Jemielity, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1954546 Å) | | Cite: | Novel N7-Arylmethyl Substituted Dinucleotide mRNA 5' cap Analogs: Synthesis and Evaluation as Modulators of Translation.
Pharmaceutics, 13, 2021
|
|
6AQX
 
 | | Crystal Structure of Z-DNA with 6-fold Twinning_Z4B | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Luo, Z, Dauter, Z, Gilski, M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Four highly pseudosymmetric and/or twinned structures of d(CGCGCG)2 extend the repertoire of crystal structures of Z-DNA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5U3V
 
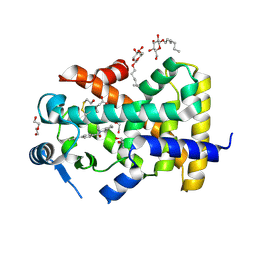 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 6 | | Descriptor: | 6-[2-({cyclopentyl[4-(furan-3-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6PJD
 
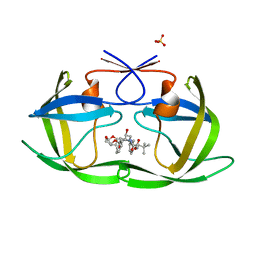 | | HIV-1 Protease NL4-3 WT in Complex with LR2-32 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-3-methyl-L-valyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6PJM
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-35 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3S,5S)-3-hydroxy-5-{[N-(methoxycarbonyl)-3-methyl-L-valyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
3VQF
 
 | | Crystal Structure Analysis of the PDZ Domain Derived from the Tight Junction Regulating Protein | | Descriptor: | E3 ubiquitin-protein ligase LNX | | Authors: | Akiyoshi, Y, Hamada, D, Goda, N, Tenno, T, Narita, H, Nakagawa, A, Furuse, M, Suzuki, M, Hiroaki, H. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural basis for down regulation of tight junction by PDZ-domain containing E3-Ubiquitin ligase
To be Published
|
|
7JY2
 
 | | Z-DNA joint X-ray/Neutron | | Descriptor: | Chains: A,B | | Authors: | Harp, J.M, Coates, L, Egli, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | Water structure around a left-handed Z-DNA fragment analyzed by cryo neutron crystallography.
Nucleic Acids Res., 49, 2021
|
|
7MU4
 
 | | Crystal Structure of HPV L1-directed D24.M01Fab | | Descriptor: | D24.M01 Fab Heavy Chain, D24.M01 Fab Light Chain, DI(HYDROXYETHYL)ETHER | | Authors: | Singh, S, Pancera, M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterisation of Immune Responses to HPV Vaccination
To Be Published
|
|
5UDV
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with iron | | Descriptor: | FE (III) ION, Lactate racemization operon protein LarE, PHOSPHATE ION, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2MN3
 
 | | Structure of Platypus 'Intermediate' Defensin-like Peptide (Int-DLP) | | Descriptor: | Defensin-BvL | | Authors: | Torres, A.M, Bansal, P.S, Koh, J.M.S, Pages, G, Wu, M.J, Kuchel, P.W. | | Deposit date: | 2014-03-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and antimicrobial activity of platypus 'intermediate' defensin-like peptide.
Febs Lett., 588, 2014
|
|
3VVL
 
 | | Crystal structure of L-serine-O-acetyltransferase found in D-cycloserine biosynthetic pathway | | Descriptor: | Homoserine O-acetyltransferase | | Authors: | Oda, K, Matoba, Y, Kumagai, T, Noda, M, Sugiyama, M. | | Deposit date: | 2012-07-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic study to determine the substrate specificity of an L-serine-acetylating enzyme found in the D-cycloserine biosynthetic pathway
J.Bacteriol., 195, 2013
|
|
6YRC
 
 | | Spectroscopically-validated structure of DtpB from Streptomyces lividans in the ferric state | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lucic, M, Dworkowski, F.S.N, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2020-04-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Serial Femtosecond Zero Dose Crystallography Captures a Water-Free Distal Heme Site in a Dye-Decolorising Peroxidase to Reveal a Catalytic Role for an Arginine in Fe IV =O Formation.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7MO1
 
 | | Crystal Structure of the ZnF1 of Nucleoporin NUP153 in complex with Ran-GDP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNW
 
 | | Crystal Structure of Nup358/RanBP2 Ran-binding domain 1 in complex with Ran-GPPNHP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNZ
 
 | | Crystal Structure of Nup358/RanBP2 Ran-binding domain 4 in complex with Ran-GPPNHP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
