6H68
 
 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) with fully-ordered A49 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6WQ3
 
 | | Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 in Complex with 7-methyl-GpppA and S-adenosyl-L-homocysteine. | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
4K48
 
 | | Structure of the Streptococcus pneumoniae leucyl-tRNA synthetase editing domain | | Descriptor: | Leucine--tRNA ligase | | Authors: | Hu, Q.H, Liu, R.J, Fang, Z.P, Zhang, J, Ding, Y.Y, Tan, M, Wang, M, Pan, W, Zhou, H.C, Wang, E.D. | | Deposit date: | 2013-04-12 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery of a potent benzoxaborole-based anti-pneumococcal agent targeting leucyl-tRNA synthetase
Sci Rep, 3, 2013
|
|
4K6A
 
 | | Revised Crystal Structure of apo-form of Triosephosphate Isomerase (tpiA) from Escherichia coli at 1.8 Angstrom Resolution. | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Minasov, G, Kuhn, M, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
PLoS ONE, 9, 2014
|
|
1JKB
 
 | | HUMAN LYSOZYME MUTANT WITH GLU 35 REPLACED BY ALA | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Muraki, M, Harata, K, Goda, S, Nagahora, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Importance of van der Waals contact between Glu 35 and Trp 109 to the catalytic action of human lysozyme.
Protein Sci., 6, 1997
|
|
6QQL
 
 | | Crystal structure of Porphyromonas gingivalis glutaminyl cyclase | | Descriptor: | Glutamine cyclotransferase, ZINC ION | | Authors: | Linnert, M, Piechotta, A, Parthier, C, Taudte, N, Kolenko, P, Rahfeld, J, Potempa, J, Stubbs, M.T. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.814 Å) | | Cite: | Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis.
J.Biol.Chem., 296, 2021
|
|
4Q24
 
 | | Crystal structure of Cyclo(L-leucyl-L-phenylalanyl) synthase | | Descriptor: | Cyclo(L-leucyl-L-phenylalanyl) synthase, PHENYLMETHYL N-[(2S)-4-CHLORO-3-OXO-1-PHENYL-BUTAN-2-YL]CARBAMATE | | Authors: | Moutiez, M, Schmitt, E, Seguin, J, Thai, R, Favry, E, Mechulam, Y, Gondry, M. | | Deposit date: | 2014-04-07 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unravelling the mechanism of non-ribosomal peptide synthesis by cyclodipeptide synthases.
Nat Commun, 5, 2014
|
|
2YM6
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2R)-4-(9H-pyrido[4',3':4,5]pyrrolo[2,3-d]pyrimidin-4-yl)morpholin-2-yl]methanamine, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
2R2Z
 
 | | The crystal structure of a hemolysin domain from Enterococcus faecalis V583 | | Descriptor: | Hemolysin, ZINC ION | | Authors: | Zhang, R, Tan, K, Zhou, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of a hemolysin domain from Enterococcus faecalis V583.
To be Published
|
|
5D91
 
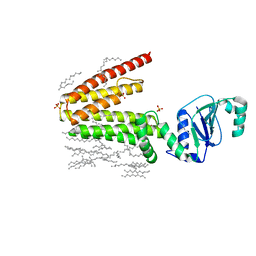 | | Structure of a phosphatidylinositolphosphate (PIP) synthase from Renibacterium Salmoninarum | | Descriptor: | AF2299 protein,Phosphatidylinositol synthase, MAGNESIUM ION, Octadecane, ... | | Authors: | Clarke, O.B, Tomasek, D.T, Jorge, C.D, Belcher Dufrisne, M, Kim, M, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Santos, H, Mancia, F. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for phosphatidylinositol-phosphate biosynthesis.
Nat Commun, 6, 2015
|
|
1JNN
 
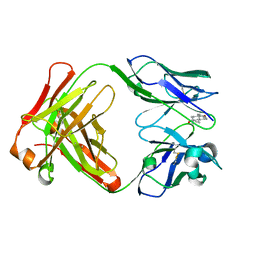 | | Crystal Structure of Fab-Estradiol Complexes | | Descriptor: | ESTRADIOL, MONOCLONAL ANTI-ESTRADIOL 17E12E5 IMMUNOGLOBULIN GAMMA-1 CHAIN, MONOCLONAL ANTI-ESTRADIOL 17E12E5 IMMUNOGLOBULIN KAPPA CHAIN | | Authors: | Monnet, C, Bettsworth, F, Stura, E.A, Le Du, M.-H, Menez, R, Derrien, L, Zinn-Justin, S, Gilquin, B, Sibai, G, Battail-Poirot, N, Jolivet, M, Menez, A, Arnaud, M, Ducancel, F, Charbonnier, J.B. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Highly specific anti-estradiol antibodies: structural characterisation and binding diversity.
J.Mol.Biol., 315, 2002
|
|
6QSX
 
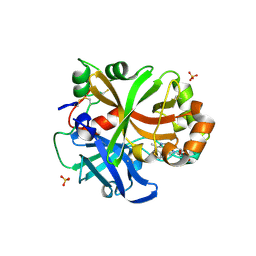 | | Complement factor B protease domain in complex with the reversible inhibitor ((2S,4S)-1-((5,7-dimethyl-1H-indol-4-yl)methyl)-4-methoxypiperidin-2-yl)methanol. | | Descriptor: | Complement factor B, SULFATE ION, ZINC ION, ... | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6H1N
 
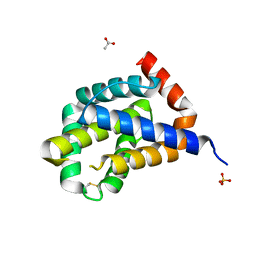 | |
1JNH
 
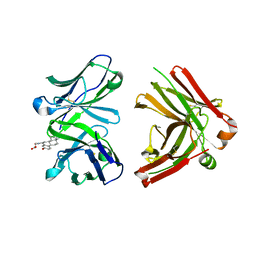 | | Crystal Structure of Fab-Estradiol Complexes | | Descriptor: | ESTRADIOL-6 CARBOXYL-METHYL-OXIME, monoclonal anti-estradiol 10G6D6 Fab heavy chain, monoclonal anti-estradiol 10G6D6 Fab light chain | | Authors: | Monnet, C, Bettsworth, F, Stura, E.A, Le Du, M.-H, Menez, R, Derrien, L, Zinn-Justin, S, Gilquin, B, Sibai, G, Battail-Poirot, N, Jolivet, M, Menez, A, Arnaud, M, Ducancel, F, Charbonnier, J.B. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Highly specific anti-estradiol antibodies: structural characterisation and binding diversity.
J.Mol.Biol., 315, 2002
|
|
1JU8
 
 | | Solution structure of Leginsulin, a plant hormon | | Descriptor: | Leginsulin | | Authors: | Yamazaki, T, Takaoka, M, Katoh, E, Hanada, K, Sakita, M, Sakata, K, Nishiuchi, Y, Hirano, H. | | Deposit date: | 2001-08-23 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A possible physiological function and the tertiary structure of a 4-kDa peptide in legumes
EUR.J.BIOCHEM., 270, 2003
|
|
2YM3
 
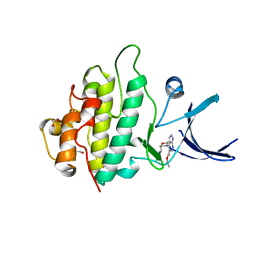 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, ETHYL 4-(2-(AMINOMETHYL)MORPHOLINO)-1H-PYRAZOLO[3,4-B]PYRIDINE-5-CARBOXYLATE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
3WGB
 
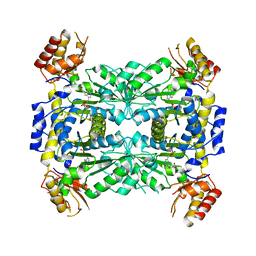 | | Crystal structure of aeromonas jandaei L-allo-threonine aldolase | | Descriptor: | GLYCINE, L-allo-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Ohtsuka, J, Hou, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | L-allo-Threonine aldolase with an H128Y/S292R mutation from Aeromonas jandaei DK-39 reveals the structural basis of changes in substrate stereoselectivity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2QGM
 
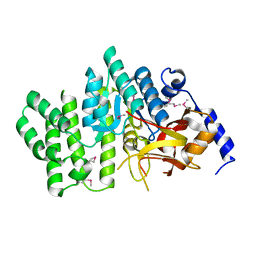 | | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136. | | Descriptor: | Succinoglycan biosynthesis protein | | Authors: | Kuzin, A.P, Abashidze, M, Jayaraman, S, Wang, H, Fang, Y, Maglaqui, M, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136.
To be Published
|
|
2YM8
 
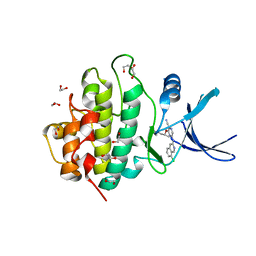 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | (R)-5-(8-CHLOROISOQUINOLIN-3-YLAMINO)-3-(1-(DIMETHYLAMINO)PROPAN-2-YLOXY)PYRAZINE-2-CARBONITRILE, 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
2QJG
 
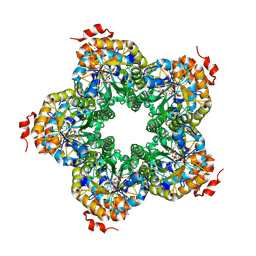 | | M. jannaschii ADH synthase complexed with F1,6P | | Descriptor: | 1,6-DI-O-PHOSPHONO-D-ALLITOL, Putative aldolase MJ0400 | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
Biochemistry, 46, 2007
|
|
7CSZ
 
 | | Crystal structure of the N-terminal tandem RRM domains of RBM45 in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*GP*GP*GP*AP*CP*GP*C)-3'), RNA-binding protein 45 | | Authors: | Chen, X, Yang, Z, Wang, W, Wang, M. | | Deposit date: | 2020-08-17 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for RNA recognition by the N-terminal tandem RRM domains of human RBM45.
Nucleic Acids Res., 49, 2021
|
|
5O77
 
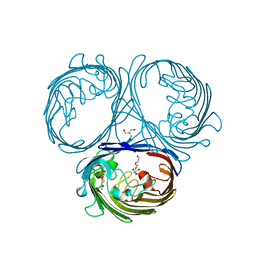 | | Klebsiella pneumoniae OmpK35 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, OmpK35 | | Authors: | van den berg, B, Pathania, M, Zahn, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
6QUS
 
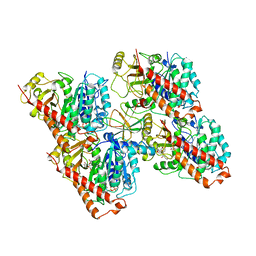 | | HsCKK (human CAMSAP1) decorated 13pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
1K23
 
 | | Inorganic Pyrophosphatase (Family II) from Bacillus subtilis | | Descriptor: | MANGANESE (II) ION, Manganese-dependent inorganic pyrophosphatase | | Authors: | Ahn, S, Milner, A.J, Futterer, K, Konopka, M, Ilias, M, Young, T.W, White, S.A. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The "open" and "closed" structures of the type-C inorganic pyrophosphatases from Bacillus subtilis and Streptococcus gordonii.
J.Mol.Biol., 313, 2001
|
|
6H64
 
 | | Crystal structure of the CRD-SAT | | Descriptor: | Galectin-3, SULFATE ION, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Charron, C, Kriznik, A, Yelehe-Okouma, M, Jouzeau, J.-Y, Reboul, P. | | Deposit date: | 2018-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRD SAT Generated by pCARGHO: A New Efficient Lectin-Based Affinity Tag Method for Safe, Simple, and Low-Cost Protein Purification.
Biotechnol J, 14, 2019
|
|
