6YPK
 
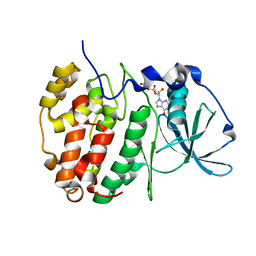 | | Crystal Structure of CK2alpha with GTP bound | | Descriptor: | Casein kinase II subunit alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2020-04-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Proposed Allosteric Inhibitors Bind to the ATP Site of CK2 alpha.
J.Med.Chem., 63, 2020
|
|
6YIH
 
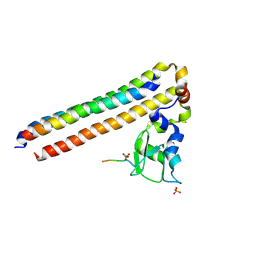 | | Structure of Chromosomal Passenger Complex (CPC) bound to phosphorylated Histone 3 peptide at 2.6 A. | | Descriptor: | Baculoviral IAP repeat-containing protein 5, Borealin, Histone H3.1, ... | | Authors: | Serena, M, Elliott, P.R, Barr, F.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle.
J.Cell Biol., 219, 2020
|
|
6YFG
 
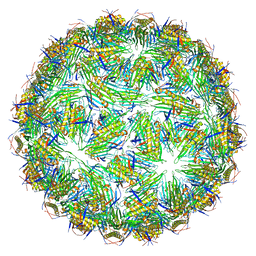 | | Virus-like particle of Beihai levi-like virus 32 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.897 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YQ2
 
 | |
6YQI
 
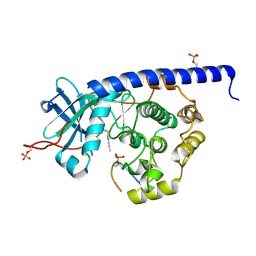 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with long-chain Fasudil-derivative N-[2-(propylamino)ethyl]isoquinoline-5-sulfonamide (soaked) | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ~{N}-[2-(propylamino)ethyl]isoquinoline-5-sulfonamide | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-04-17 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6YPY
 
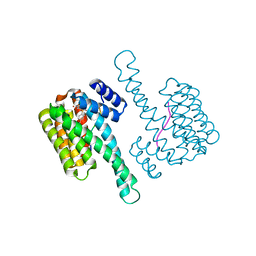 | |
6YHU
 
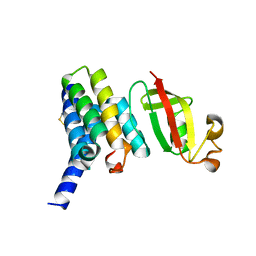 | |
6YFL
 
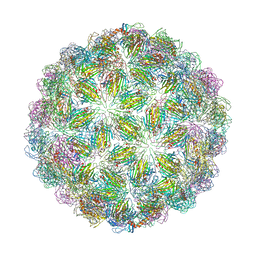 | | Virus-like particle of bacteriophage ESE020 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YIJ
 
 | | Crystal structure of the CREBBP bromodomain in complex with a benzo-diazepine ligand | | Descriptor: | (4~{R})-6-[(~{E})-5-(7-methoxy-3,4-dihydro-2~{H}-quinolin-1-yl)pent-1-enyl]-4-methyl-1,3,4,5-tetrahydro-1,5-benzodiazepin-2-one, CREBBP | | Authors: | Picaud, S, Brand, M, Tobias, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Conway, S, Filippakopoulos, P. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the CREBBP bromodomain in complex with a benzo-diazepine ligand
To Be Published
|
|
6YFT
 
 | | Virus-like particle of Wenzhou levi-like virus 1 | | Descriptor: | RNA, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YO4
 
 | | Para-Carborane propyl-sulfonamide in complex with CA IX mimic | | Descriptor: | Carbonic anhydrase 2, Para-Carborane propyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-04-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Direct Introduction of an Alkylsulfonamido Group on C-sites of Isomeric Dicarba-closo-dodecaboranes: The Influence of Stereochemistry on Inhibitory Activity against the Cancer-Associated Carbonic Anhydrase IX Isoenzyme.
Chemistry, 26, 2020
|
|
6YOK
 
 | | Para-Carborane di-propyl-sulfonamide in complex with CA IX mimic | | Descriptor: | Carbonic anhydrase 2, Para-Carborane di-propyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-04-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Direct Introduction of an Alkylsulfonamido Group on C-sites of Isomeric Dicarba-closo-dodecaboranes: The Influence of Stereochemistry on Inhibitory Activity against the Cancer-Associated Carbonic Anhydrase IX Isoenzyme.
Chemistry, 26, 2020
|
|
6YOW
 
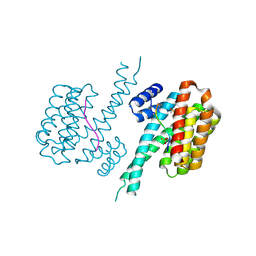 | |
6YNB
 
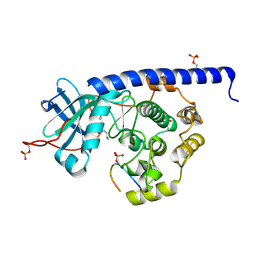 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with short-chain Fasudil-derivative N-(2-aminoethyl)isoquinoline-5-sulfonamide (soaked) | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-AMINOETHYL)ISOQUINOLINE-5-SULFONAMIDE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-04-13 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6YPJ
 
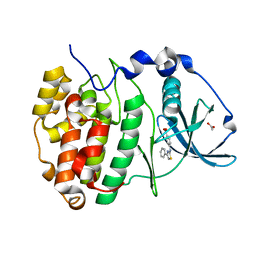 | | Crystal Structure of CK2alpha with Compound 1 bound | | Descriptor: | 4-[(4-phenyl-1,3-thiazol-2-yl)amino]benzoic acid, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2020-04-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Proposed Allosteric Inhibitors Bind to the ATP Site of CK2 alpha.
J.Med.Chem., 63, 2020
|
|
6YPL
 
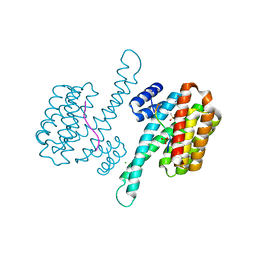 | |
6YCC
 
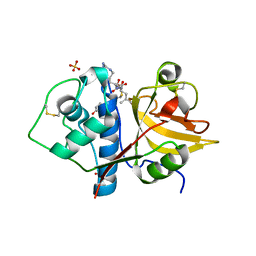 | | Structure the ananain protease from Ananas comosus covalently bound to the E64 inhibitor | | Descriptor: | Ananain, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6YFH
 
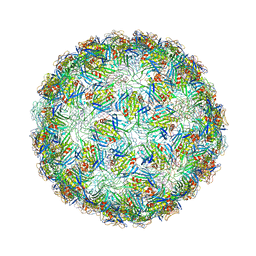 | | Virus-like particle of bacteriophage EMS014 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.893 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YDI
 
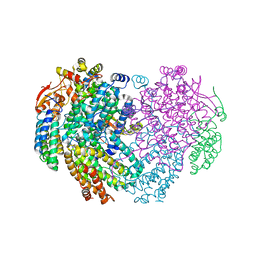 | | XFEL structure of the Soluble methane monooxygenase hydroxylase and regulatory subunit complex, from Methylosinus trichosporium OB3b, diferrous state | | Descriptor: | FE (II) ION, GLYCEROL, Methane monooxygenase, ... | | Authors: | Srinivas, V, Hogbom, M. | | Deposit date: | 2020-03-20 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-Resolution XFEL Structure of the Soluble Methane Monooxygenase Hydroxylase Complex with its Regulatory Component at Ambient Temperature in Two Oxidation States.
J. Am. Chem. Soc., 142, 2020
|
|
6YFQ
 
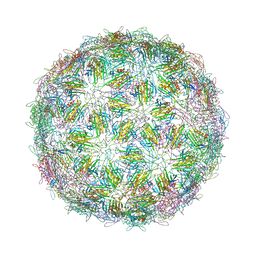 | | Virus-like particle of bacteriophage NT-214 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFA
 
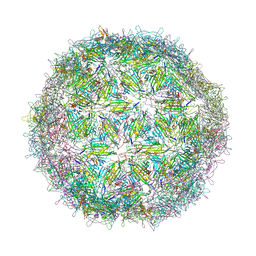 | | Virus-like particle of bacteriophage AVE015 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFN
 
 | | Virus-like particle of bacteriophage ESE058 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.189 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YIL
 
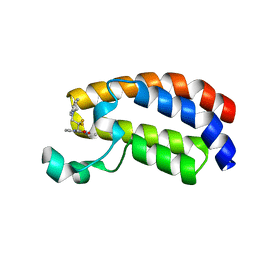 | | Crystal structure of the CREBBP bromodomain in complex with a tetrahydroquinoxaline ligand | | Descriptor: | (3~{R})-~{N}-[3-(3,4-dihydro-2~{H}-quinolin-1-yl)-2,2-bis(fluoranyl)propyl]-3-methyl-2-oxidanylidene-3,4-dihydro-1~{H}-quinoxaline-5-carboxamide, CREBBP | | Authors: | Picaud, S, Brand, M, Tobias, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Conway, S, Filippakopoulos, P. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of the CREBBP bromodomain in complex with a tetrahydroquinoxaline ligand
To Be Published
|
|
6YNR
 
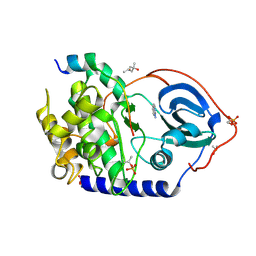 | | Crystal structure of the cAMP-dependent protein kinase A in complex with 1,7-Naphthyridin-8-amine (soaked) and PKI (5-24) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,7-naphthyridin-8-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Oebbeke, M, Heine, A, Klebe, G. | | Deposit date: | 2020-04-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6YIP
 
 | | Structure of MKLP2 coiled coil | | Descriptor: | ISOPROPYL ALCOHOL, Kinesin-like protein KIF20A | | Authors: | Serena, M, Elliott, P.R, Barr, F.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle.
J.Cell Biol., 219, 2020
|
|
