8WDQ
 
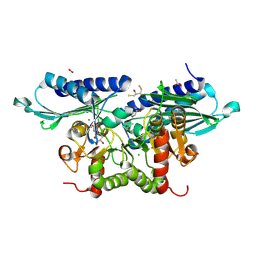 | | Crystal Structure of Pseudomonas aeruginosa SuhB in complex with D-myo-inositol-1-phosphate | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yadav, V.K, Maji, S, Shukla, M, Bhattacharyya, S. | | Deposit date: | 2023-09-15 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Pseudomonas aeruginosa SuhB in complex with D-myo-inositol-1-phosphate
To Be Published
|
|
7P5V
 
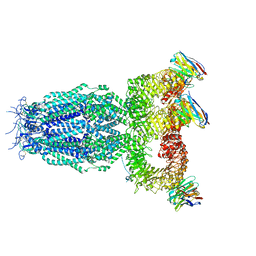 | |
7P60
 
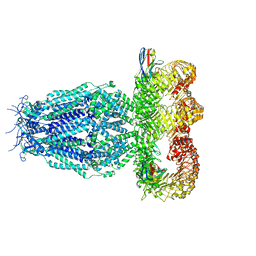 | |
2CLX
 
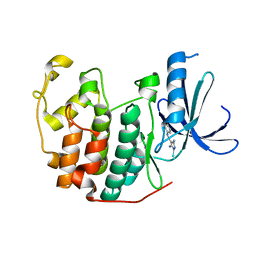 | | 4-Arylazo-3,5-diamino-1H-pyrazole CDK Inhibitors: SAR Study, Crystal Structure in Complex with CDK2, Selectivity, and Cellular Effects | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Krystof, V, Cankar, P, Frysova, I, Slouka, J, Kontopidis, G, Dzubak, P, Hajduch, M, Deazevedo, W.F, Paprskarova, M, Orsag, M, Rolcik, J, Latr, A, Fischer, P.M, Strnad, M. | | Deposit date: | 2006-05-02 | | Release date: | 2006-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 4-Arylazo-3,5-Diamino-1H-Pyrazole Cdk Inhibitors: Sar Study, Crystal Structure in Complex with Cdk2, Selectivity, and Cellular Effects
J.Med.Chem., 49, 2006
|
|
8WJF
 
 | |
7OKW
 
 | | 1.62A X-ray crystal structure of the conserved C-terminal (CCT) of human OSR1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Bax, B.D, Elvers, K.T, Lipka-Lloyd, M, Mehellou, Y. | | Deposit date: | 2021-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of the Human SPAK and OSR1 Conserved C-Terminal (CCT) Domains.
Chembiochem, 23, 2022
|
|
8WM1
 
 | | DHS dehydratase | | Descriptor: | 3-dehydroshikimate dehydratase (DHS dehydratase), CALCIUM ION | | Authors: | Wang, M. | | Deposit date: | 2023-10-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | DHS dehydratase
To Be Published
|
|
8WIP
 
 | |
8WFM
 
 | |
6QGY
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody B12 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoB12, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
8WIE
 
 | |
7P5Y
 
 | |
8WG3
 
 | | mouse TMEM63b in LMNG-CHS micelle | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
8WFH
 
 | |
7P5W
 
 | |
7UJV
 
 | | Structure of PHD2 in complex with HIF2a-CODD | | Descriptor: | Egl nine homolog 1, Endothelial PAS domain-containing protein 1, FE (III) ION, ... | | Authors: | Ferens, F.G, Tarade, D, Lee, J.E, Ohh, M. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deficiency in PHD2-mediated hydroxylation of HIF2 alpha underlies Pacak-Zhuang syndrome.
Commun Biol, 7, 2024
|
|
8WG4
 
 | | mouse TMEM63b in DDM-CHS micelle with YN9303-24 Fab | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
7P6K
 
 | |
6I94
 
 | |
8WIQ
 
 | |
8WKW
 
 | | Structure of MAVS-CARD Filament | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Shi, M, Gao, P. | | Deposit date: | 2023-09-28 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure of MAVS-CARD Filament
To Be Published
|
|
7EY3
 
 | | Double cysteine mutations in T1 lipase | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Hamdan, S.H, Leow, T.C, Yahaya, N.M, Ali, M.S.M, Jonet, M.A, Mohamad Aris, S.N.A, Maiangwa, J. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Knotting terminal ends of mutant T1 lipase with disulfide bond improved structure rigidity and stability.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
7XEB
 
 | | Collagenase from Grimontia (Vibrio) hollisae 1706B complexed with Gly-Pro-Hyp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLY-PRO-HYP peptide, ... | | Authors: | Ikeuchi, T, Yasumoto, M, Takita, T, Mizutani, K, Mikami, B, Tanaka, K, Hattori, S, Yasukawa, K. | | Deposit date: | 2022-03-30 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of Grimontia hollisae collagenase provides insights into its novel substrate specificity toward collagen.
J.Biol.Chem., 298, 2022
|
|
8WOK
 
 | |
7OSB
 
 | | Crystal Structure of a Double Mutant PETase (S238F/W159H) from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ethylene terephthalate) hydrolase, ... | | Authors: | Shakespeare, T.J, Zahn, M, Allen, M.D, McGeehan, J.E. | | Deposit date: | 2021-06-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Comparative Performance of PETase as a Function of Reaction Conditions, Substrate Properties, and Product Accumulation.
ChemSusChem, 15, 2022
|
|
