7R1G
 
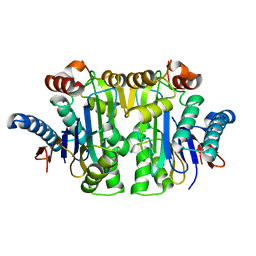 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-38 (R207C, D210S, S211V) | | Descriptor: | Beta-aspartyl-peptidase, Isoaspartyl peptidase, SODIUM ION | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5GUL
 
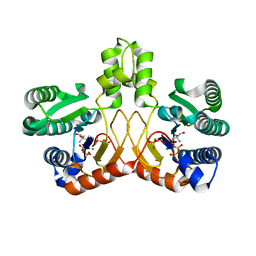 | | Crystal structure of Tris/PPix2/Mg2+ bound form of cyclolavandulyl diphosphate synthase (CLDS) from Streptomyces sp. CL190 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cyclolavandulyl diphosphate synthase, MAGNESIUM ION, ... | | Authors: | Tomita, T, Kobayashi, M, Nishiyama, M, Kuzuyama, T. | | Deposit date: | 2016-08-29 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure and Mechanism of the Monoterpene Cyclolavandulyl Diphosphate Synthase that Catalyzes Consecutive Condensation and Cyclization.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MYS
 
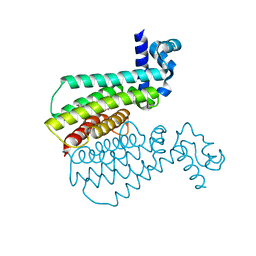 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK920684A at 1.59A resolution | | Descriptor: | 2-(3-fluoranylphenoxy)-~{N}-(4-pyridin-2-yl-1,3-thiazol-2-yl)ethanamide, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
8K0D
 
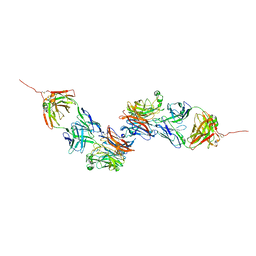 | |
1VTJ
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 1 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1999-09-14 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Structure of the Netropsin-d(CGCGATATCGCG) Complex: DNA Conformation in an Alternating AT Segment
Biochemistry, 28, 1989
|
|
5H0E
 
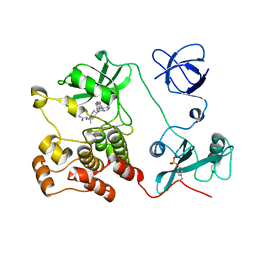 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-4-methyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
1LVA
 
 | |
4YM8
 
 | |
1VDS
 
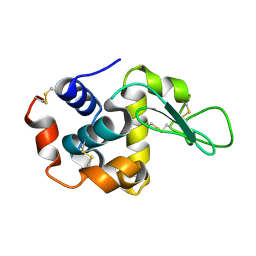 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space
to be published
|
|
7C38
 
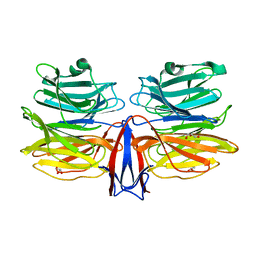 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with L-fucose | | Descriptor: | AofleA, GLYCEROL, alpha-L-fucopyranose, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
5H0G
 
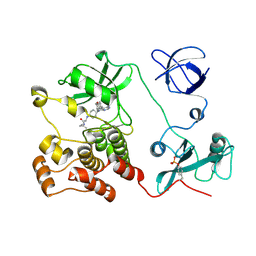 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-N,4-dimethylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-~{N},4-dimethyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
6WOR
 
 | |
4OYM
 
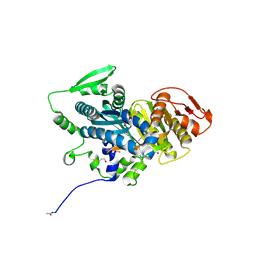 | | Human solAC Complexed with (4-Amino-furazan-3-yl)-(3-methoxy-phenyl)-methanone | | Descriptor: | (4-azanyl-1,2,5-oxadiazol-3-yl)-(3-methoxyphenyl)methanone, Adenylate cyclase type 10, GLYCEROL | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-12 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
4OYX
 
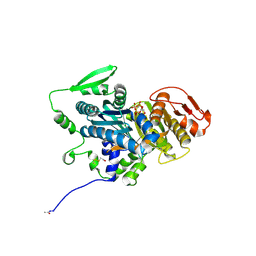 | | Human solAC Complexed with AMPCPP | | Descriptor: | Adenylate cyclase type 10, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
3KZZ
 
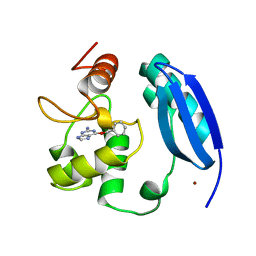 | |
7RN2
 
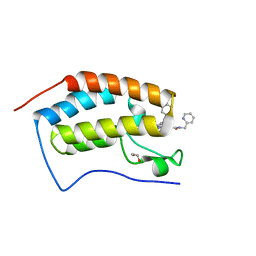 | | Crystal structure of the first bromodomain of human BRD4 in complex with SJ001010551-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(6S,10S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-[(pyridin-2-yl)methyl]acetamide, Bromodomain-containing protein 4 | | Authors: | Stachowski, T.R, Fischer, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | From PROTAC to inhibitor: Structure-guided discovery of potent and orally bioavailable BET inhibitors.
Eur.J.Med.Chem., 251, 2023
|
|
4OIZ
 
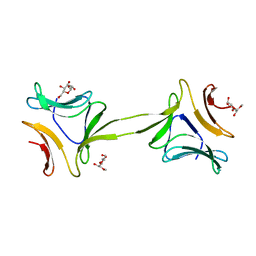 | | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis | | Descriptor: | LysM domain protein, methyl alpha-D-mannopyranoside | | Authors: | Patra, D, Mishra, P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-01-20 | | Release date: | 2014-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis.
Glycobiology, 24, 2014
|
|
7RMD
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SJ001011461-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(6S,10R)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(1,3,4-thiadiazol-2-yl)acetamide, Bromodomain-containing protein 4 | | Authors: | Stachowski, T.R, Fischer, M. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | From PROTAC to inhibitor: Structure-guided discovery of potent and orally bioavailable BET inhibitors.
Eur.J.Med.Chem., 251, 2023
|
|
2L7H
 
 | |
4YND
 
 | | The Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2 | | Descriptor: | N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[(2R)-2-hydroxy-2-(3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Sweis, R.F, Wang, Z, Algire, M, Arrowsmith, C.H, Brown, P.J, Chiang, G.C, Guo, J, Jakob, C.G, Kennedy, S, Li, F, Soni, N.B, Vedadi, M, Pappano, W.N. | | Deposit date: | 2015-03-09 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2.
Acs Med.Chem.Lett., 6, 2015
|
|
8JRD
 
 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waki, T, Imaizumi, R, Nakata, S, Yanai, T, Takeshita, K, Sakai, N, Kataoka, K, Yamamoto, M, Nakayama, T, Yamashita, S. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into catalytic promiscuity of chalcone synthase from Glycine max (L.) Merr.: Coenzyme A-induced alteration of product specificity.
Biochem.Biophys.Res.Commun., 718, 2024
|
|
7C3E
 
 | | Crystal structure of R97A/R150A/R203A mutant of AofleA from Arthrobotrys oligospora | | Descriptor: | AofleA, GLYCEROL, SULFATE ION | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
1VDQ
 
 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution
to be published
|
|
5MYP
 
 | | Structure of apo-TbALDH3 | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL | | Authors: | Zoltner, M, Zhang, N, Horn, D, Field, M.C. | | Deposit date: | 2017-01-27 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Activation of an anti-trypanosomal benzoxaborole requires both host and parasite factors
To Be Published
|
|
1VDT
 
 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.7 angstroms resolution under basic conditions in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.7 angstroms resolution under basic conditions in space
to be published
|
|
