7Y4A
 
 | | Crystal structure of human ELMO1 RBD-RhoG complex | | Descriptor: | Engulfment and cell motility protein 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tsuda, K, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Targeting Ras-binding domain of ELMO1 by computational nanobody design.
Commun Biol, 6, 2023
|
|
8J26
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-4, AM047-6, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
8IYJ
 
 | | Cryo-EM structure of the 48-nm repeat doublet microtubule from mouse sperm | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Zhou, L.N, Gui, M, Wu, J.P. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of sperm flagellar doublet microtubules expand the genetic spectrum of male infertility.
Cell, 186, 2023
|
|
8J1Q
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-0, AM047-0, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
8J07
 
 | |
5KA8
 
 | | Protein Tyrosine Phosphatase 1B L192A mutant, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
6APM
 
 | | Hen egg-white lysozyme (WT), solved with serial millisecond crystallography using synchrotron radiation | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Lyubimov, A.Y, Mathews, I.I, Uervivojnangkoorn, M, Soltis, S.M, Cohen, A.E. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
6ND5
 
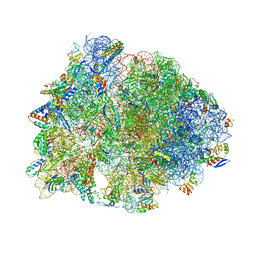 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with chloramphenicol and bound to mRNA and A-, P-, and E-site tRNAs at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
8J62
 
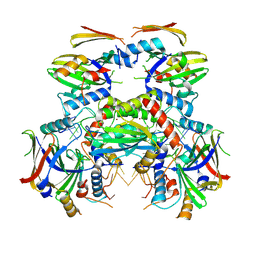 | | Cryo-EM structure of APOBEC3G-Vif complex | | Descriptor: | APOBEC3G, Core binding factor beta, RNA (5'-R(*CP*GP*GP*UP*UP*GP*AP*UP*UP*GP*UP*UP*UP*UP*AP*AP*CP*AP*A)-3'), ... | | Authors: | Kouno, T, Shibata, S, Hyun, J, Kim, T.G, Wolf, M. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into RNA bridging between HIV-1 Vif and antiviral factor APOBEC3G.
Nat Commun, 14, 2023
|
|
3CU8
 
 | | Impaired binding of 14-3-3 to Raf1 is linked to Noonan and LEOPARD syndrome | | Descriptor: | 14-3-3 protein zeta/delta, MAGNESIUM ION, PROPANOIC ACID, ... | | Authors: | Schumacher, B, Weyand, M, Kuhlmann, J, Ottmann, C. | | Deposit date: | 2008-04-16 | | Release date: | 2009-05-05 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling.
Mol. Cell. Biol., 30, 2010
|
|
6NDY
 
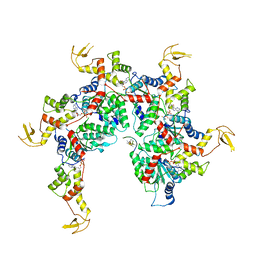 | | Vps4 with Cyclic Peptide Bound in the Central Pore | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Designed Cyclic Peptide, ... | | Authors: | Han, H, Fulcher, J.M, Dandey, V.P, Sundquist, W.I, Kay, M.S, Shen, P, Hill, C.P. | | Deposit date: | 2018-12-14 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases.
Elife, 8, 2019
|
|
3QZZ
 
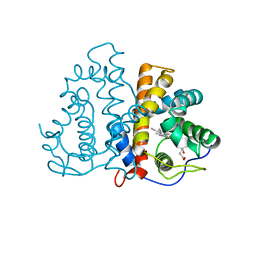 | | 3D Structure of Ferric Methanosarcina Acetivorans Protoglobin Y61W mutant in Aquomet form | | Descriptor: | Methanosarcina acetivorans protoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Tilleman, L, Dewilde, S, Ascenzi, P, Coletta, M, Ciaccio, C, Bruno, S, Moens, L, Bolognesi, M, Nardini, M. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural heterogeneity and ligand gating in ferric methanosarcina acetivorans protoglobin mutants.
Iubmb Life, 63, 2011
|
|
5JPF
 
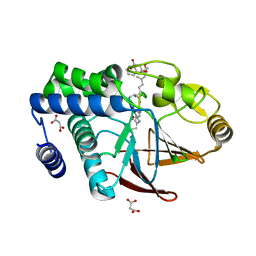 | | Serine/Threonine phosphatase Z1 (Candida albicans) binds to inhibitor microcystin-LR | | Descriptor: | MALONATE ION, MANGANESE (II) ION, Microcystin-LR, ... | | Authors: | Choy, M.S, Chen, E.H, Peti, W, Page, R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.39929724 Å) | | Cite: | Molecular Insights into the Fungus-Specific Serine/Threonine Protein Phosphatase Z1 in Candida albicans.
Mbio, 7, 2016
|
|
5XQJ
 
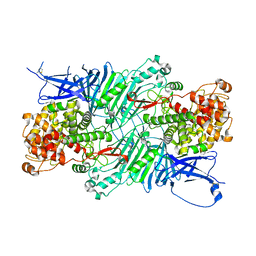 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-[beta-D-galactopyranose-(1-4)]alpha-L-rhamnopyranose, 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, ... | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
6NCK
 
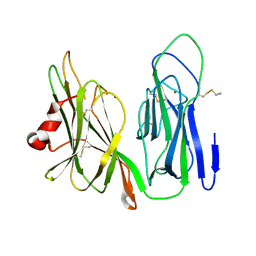 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | COPPER (II) ION, NICKEL (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Miller, M.S, Maheshwari, S, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
4PH4
 
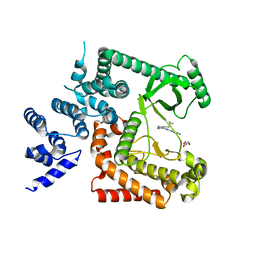 | | The crystal structure of Human VPS34 in complex with PIK-III | | Descriptor: | 4'-(cyclopropylmethyl)-N~2~-(pyridin-4-yl)-4,5'-bipyrimidine-2,2'-diamine, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3 | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2014-05-04 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo.
Nat.Cell Biol., 16, 2014
|
|
7E18
 
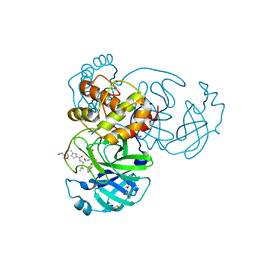 | | Crystal structure of SAR-CoV-2 3CL protease complex with inhibitor YH-53 | | Descriptor: | 1,2-ETHANEDIOL, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1ab | | Authors: | Senda, M, Konno, S, Hayashi, Y, Senda, T. | | Deposit date: | 2021-02-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents.
J.Med.Chem., 65, 2022
|
|
7E19
 
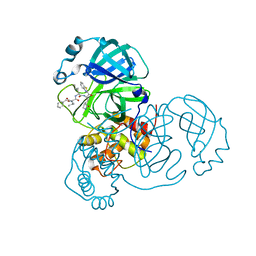 | | Crystal structure of SAR-CoV-2 3CL protease complex with inhibitor SH-5 | | Descriptor: | (phenylmethyl) N-[(2S)-1-[[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]carbamate, 3C-like proteinase | | Authors: | Senda, M, Konno, S, Hayashi, Y, Senda, T. | | Deposit date: | 2021-02-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents.
J.Med.Chem., 65, 2022
|
|
5LV9
 
 | |
8JH0
 
 | | Crystal structure of the light-driven sodium pump IaNaR | | Descriptor: | RETINAL, Xanthorhodopsin | | Authors: | Hashimoto, T, Kato, K, Tanaka, Y, Yao, M, Kikukawa, T. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Multistep conformational changes leading to the gate opening of light-driven sodium pump rhodopsin.
J.Biol.Chem., 299, 2023
|
|
5ZA7
 
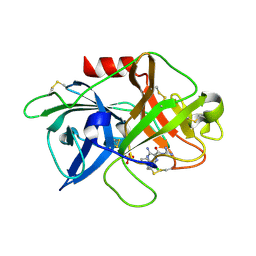 | | uPA-HMA | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-[bis(azanyl)methylidene]-6-chloranyl-pyrazine-2-carboxamide, SULFATE ION, Urokinase-type plasminogen activator chain B | | Authors: | Buckley, B.J, Jiang, L.G, Huang, M.D, Kelso, M.J, Ranson, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 6-Substituted Hexamethylene Amiloride (HMA) Derivatives as Potent and Selective Inhibitors of the Human Urokinase Plasminogen Activator for Use in Cancer.
J. Med. Chem., 61, 2018
|
|
5ZAE
 
 | | uPA-6F-HMA | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(furan-2-yl)pyrazine-2-carboxamide, SULFATE ION, Urokinase-type plasminogen activator chain B | | Authors: | Buckley, B.J, Jiang, L.G, Huang, M.D, Kelso, M.J, Ranson, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 6-Substituted Hexamethylene Amiloride (HMA) Derivatives as Potent and Selective Inhibitors of the Human Urokinase Plasminogen Activator for Use in Cancer.
J. Med. Chem., 61, 2018
|
|
2VF7
 
 | | Crystal structure of UvrA2 from Deinococcus radiodurans | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EXCINUCLEASE ABC, SUBUNIT A., ... | | Authors: | Timmins, J, Gordon, E, Caria, S, Leonard, G, Kuo, M.S, Monchois, V, McSweeney, S. | | Deposit date: | 2007-10-31 | | Release date: | 2008-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of Deinococcus radiodurans UvrA2 provide insight into DNA binding and damage recognition by UvrAs.
Structure, 17, 2009
|
|
5LNE
 
 | | E. coli F9 pilus adhesin FmlH bound to the Thomsen-Friedenreich (TF) antigen | | Descriptor: | NICKEL (II) ION, Putative Fml fimbrial adhesin FmlD, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Ruer, S, Conover, M.S, Kalas, V, Taganna, J, De Greve, H, Pinkner, J.S, Dodson, K.W, Hultgren, S.J, Remaut, H. | | Deposit date: | 2016-08-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inflammation-Induced Adhesin-Receptor Interaction Provides a Fitness Advantage to Uropathogenic E. coli during Chronic Infection.
Cell Host Microbe, 20, 2016
|
|
5ZC5
 
 | | uPA-NU-09F | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(4-fluoranyl-1-benzofuran-2-yl)pyrazine-2-carboxamide, Urokinase-type plasminogen activator chain B | | Authors: | Jiang, L.G, Buckley, B.J, Huang, M.D, Kelso, M.J, Ranson, M. | | Deposit date: | 2018-02-15 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 6-Substituted Hexamethylene Amiloride (HMA) Derivatives as Potent and Selective Inhibitors of the Human Urokinase Plasminogen Activator for Use in Cancer.
J. Med. Chem., 61, 2018
|
|
