3NBH
 
 | | Crystal structure of human RMI1C-RMI2 complex | | Descriptor: | RecQ-mediated genome instability protein 1, RecQ-mediated genome instability protein 2 | | Authors: | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
3NIR
 
 | | Crystal structure of small protein crambin at 0.48 A resolution | | Descriptor: | Crambin, ETHANOL | | Authors: | Schmidt, A, Teeter, M, Weckert, E, Lamzin, V.S. | | Deposit date: | 2010-06-16 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.48 Å) | | Cite: | Crystal structure of small protein crambin at 0.48 A resolution
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4KAF
 
 | | Crystal Structure of Haloalkane dehalogenase HaloTag7 at the resolution 1.5A, Northeast Structural Genomics Consortium (NESG) Target OR151 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Haloalkane dehalogenase, ... | | Authors: | Kuzin, A.P, Lew, S, Neklesa, T.K, Noblin, D, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, H, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Crews, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-22 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR151
To be Published
|
|
1ZX6
 
 | | High-resolution crystal structure of yeast Pin3 SH3 domain | | Descriptor: | Ypr154wp | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Zou, P, Song, Y.H, Wilmanns, M. | | Deposit date: | 2005-06-07 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics of yeast SH3 domains
To be Published
|
|
2XS1
 
 | | Crystal Structure of ALIX in complex with the SIVmac239 PYKEVTEDL Late Domain | | Descriptor: | GAG POLYPROTEIN, PROGRAMMED CELL DEATH 6-INTERACTING PROTEIN | | Authors: | Zhai, Q, Landesman, M, Robinson, H, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Identification and Structural Characterization of the Alix-Binding Late Domains of Sivmac239 and Sivagmtan-1.
J.Virol., 85, 2011
|
|
1ZX9
 
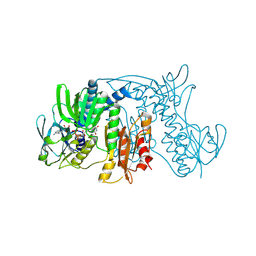 | | Crystal Structure of Tn501 MerA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mercuric reductase | | Authors: | Dong, A, Ledwidge, R, Patel, B, Fiedler, D, Falkowski, M, Zelikova, J, Summers, A.O, Pai, E.F, Miller, S.M. | | Deposit date: | 2005-06-07 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NmerA, the Metal Binding Domain of Mercuric Ion Reductase, Removes Hg(2+) from Proteins, Delivers It to the Catalytic Core, and Protects Cells under Glutathione-Depleted Conditions
Biochemistry, 44, 2005
|
|
3NJM
 
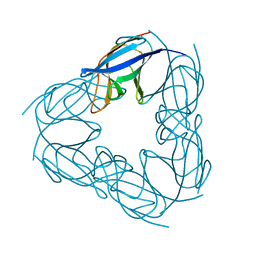 | | P117A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
1ZXK
 
 | | Crystal Structure of Cadherin8 EC1 domain | | Descriptor: | Cadherin-8 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Schieren, I, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-08 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
3NCL
 
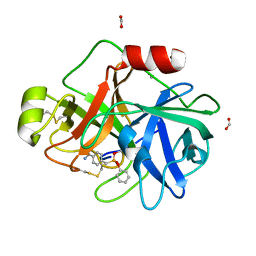 | |
4K18
 
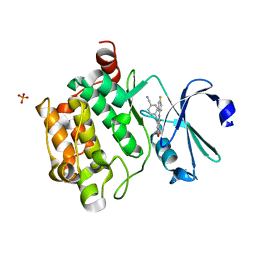 | | Structure of PIM-1 kinase bound to 5-(4-cyanobenzyl)-N-(4-fluorophenyl)-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carboxamide | | Descriptor: | 5-(4-cyanobenzyl)-N-(4-fluorophenyl)-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carboxamide, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H, Steffek, M. | | Deposit date: | 2013-04-04 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Discovery of novel pyrazolo[1,5-a]pyrimidines as potent pan-Pim inhibitors by structure- and property-based drug design.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2AEZ
 
 | | Crystal structure of fructan 1-exohydrolase IIa (E201Q) from Cichorium intybus in complex with 1-kestose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Verhaest, M, Lammens, W, Le Roy, K, De Ranter, C.J, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2005-07-25 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor.
New Phytol, 174, 2007
|
|
3NJU
 
 | | Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution | | Descriptor: | 4-METHOXYBENZOIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Kaushik, S, Prem Kumar, R, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution
To be Published
|
|
3NDI
 
 | | X-ray Structure of a C-3'-Methyltransferase in Complex with S-adenosylmethionine and dTMP | | Descriptor: | Methyltransferase, PHOSPHATE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Bruender, N.A, Thoden, J.B, Kaur, M, Avey, M.K, Holden, H.M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Architecture of a C-3'-Methyltransferase Involved in the Biosynthesis of d-Tetronitrose.
Biochemistry, 49, 2010
|
|
2XO3
 
 | |
4K08
 
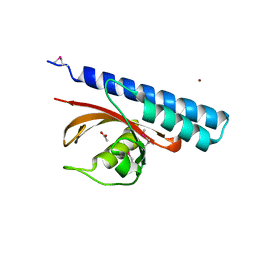 | | Periplasmic sensor domain of chemotaxis protein, Adeh_3718 | | Descriptor: | ACETATE ION, Methyl-accepting chemotaxis sensory transducer, ZINC ION | | Authors: | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
3WKV
 
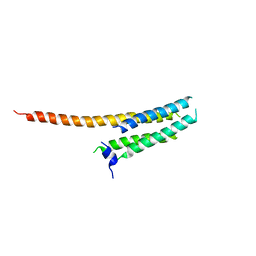 | | Voltage-gated proton channel: VSOP/Hv1 chimeric channel | | Descriptor: | Ion channel | | Authors: | Takeshita, K, Sakata, S, Yamashita, E, Fujiwara, Y, Kawanabe, A, Kurokawa, T, Okochi, Y, Matsuda, M, Narita, H, Okamura, Y, Nakagawa, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.453 Å) | | Cite: | X-ray crystal structure of voltage-gated proton channel.
Nat.Struct.Mol.Biol., 21, 2014
|
|
209D
 
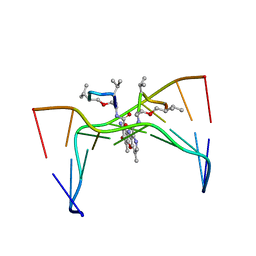 | | Structural, physical and biological characteristics of RNA:DNA binding agent N8-actinomycin D | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3'), N8-ACTINOMYCIN D | | Authors: | Shinomiya, M, Chu, W, Carlson, R.G, Weaver, R.F, Takusagawa, F. | | Deposit date: | 1995-05-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural, Physical, and Biological Characteristics of RNA.DNA Binding Agent N8-Actinomycin D.
Biochemistry, 34, 1995
|
|
1ZZE
 
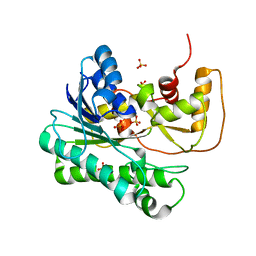 | | X-ray Structure of NADPH-dependent Carbonyl Reductase from Sporobolomyces salmonicolor | | Descriptor: | Aldehyde reductase II, SULFATE ION | | Authors: | Kamitori, S, Iguchi, A, Ohtaki, A, Yamada, M, Kita, K. | | Deposit date: | 2005-06-14 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structures of NADPH-dependent Carbonyl Reductase from Sporobolomyces salmonicolor Provide Insights into Stereoselective Reductions of Carbonyl Compounds
J.Mol.Biol., 352, 2005
|
|
2XL7
 
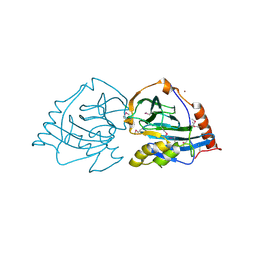 | | Structure and metal-loading of a soluble periplasm cupro-protein: Cu- CucA-closed (SeMet) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, SLL1785 PROTEIN, ... | | Authors: | Waldron, K.J, Firbank, S.J, Dainty, S.J, Perez-Rama, M, Tottey, S, Robinson, N.J. | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Metal Loading of a Soluble Periplasm Cuproprotein.
J.Biol.Chem., 285, 2010
|
|
2XPK
 
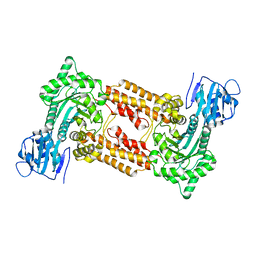 | | Cell-penetrant, nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases | | Descriptor: | N-[(5R,6R,7R,8S)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-2-(2-PHENYLETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDIN-8-YL]-3-SULFANYLPROPANAMIDE, O-GLCNACASE NAGJ | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Schimpl, M, Zheng, X, Kime, R, Read, K.D, van Aalten, D.M.F. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cell-Penetrant, Nanomolar O-Glcnacase Inhibitors Selective Against Lysosomal Hexosaminidases.
Chem.Biol, 17, 2010
|
|
213D
 
 | |
4K0Y
 
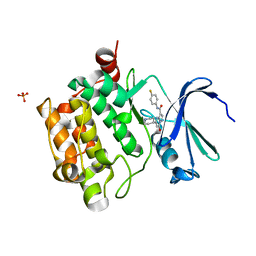 | | Structure of PIM-1 kinase bound to N-(4-fluorophenyl)-7-hydroxy-5-(piperidin-4-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Descriptor: | N-(4-fluorophenyl)-7-hydroxy-5-(piperidin-4-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H, Steffek, M. | | Deposit date: | 2013-04-04 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Discovery of novel pyrazolo[1,5-a]pyrimidines as potent pan-Pim inhibitors by structure- and property-based drug design.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3NIE
 
 | | Crystal Structure of PF11_0147 | | Descriptor: | MAP2 kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, MacKenzie, F, Kozieradzki, I, Chau, I, Lew, J, Senisterra, G, Cossar, D, Amani, M, Artz, J.D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of PF11_0147
To be Published
|
|
279D
 
 | |
2XQ1
 
 | | Crystal structure of peroxisomal catalase from the yeast Hansenula polymorpha | | Descriptor: | PEROXISOMAL CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Penya-Soler, E, Vega, M.C, Wilmanns, M, Williams, C.P. | | Deposit date: | 2010-08-31 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Features of Peroxisomal Catalase from the Yeast Hansenula Polymorpha
Acta Crystallogr.,Sect.D, 67, 2011
|
|
