3RGL
 
 | | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC in complex with ATP and glycine | | Descriptor: | (2S)-2-hydroxybutanedioic acid, ADENOSINE-5'-TRIPHOSPHATE, GLYCINE, ... | | Authors: | Tan, K, Zhang, R, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-08 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC in complex with ATP and glycine.
To be Published
|
|
4LL9
 
 | | Crystal structure of D3D4 domain of the LILRB1 molecule | | Descriptor: | IODIDE ION, Leukocyte immunoglobulin-like receptor subfamily B member 1 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
3RK8
 
 | | Crystal structure of the chloride inhibited dihydrodipicolinate synthase from Acinetobacter baumannii complexed with pyruvate at 1.8 A resolution | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dihydrodipicolinate synthase, ... | | Authors: | Kaushik, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-17 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the chloride inhibited dihydrodipicolinate synthase from Acinetobacter baumannii complexed with pyruvate at 1.8 A resolution
To be Published
|
|
4LH5
 
 | | Dual inhibition of HIV-1 replication by Integrase-LEDGF allosteric inhibitors is predominant at post-integration stage during virus production rather than at integration | | Descriptor: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, Integrase, MAGNESIUM ION | | Authors: | Ruff, M, Levy, N, Eiler, S. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage.
Retrovirology, 10, 2013
|
|
1L5H
 
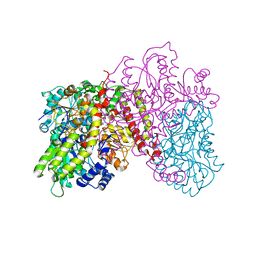 | | FeMo-cofactor Deficient Nitrogenase MoFe Protein | | Descriptor: | CALCIUM ION, FE(8)-S(7) CLUSTER, nitrogenase molybdenum-iron protein alpha chain, ... | | Authors: | Schmid, B, Ribbe, M.W, Einsle, O, Yoshida, M, Thomas, L.M, Dean, D.R, Rees, D.C, Burgess, B.K. | | Deposit date: | 2002-03-06 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a cofactor-deficient nitrogenase MoFe protein.
Science, 296, 2002
|
|
4PVN
 
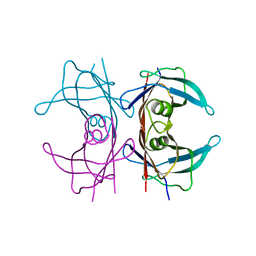 | | Neutron structure of human transthyretin (TTR) at room temperature to 2.3A resolution (monochromatic) | | Descriptor: | Transthyretin | | Authors: | Fisher, S.J, Blakeley, M.P, Haupt, M, Mason, S.A, Cooper, J.B, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-12 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (2.3 Å), X-RAY DIFFRACTION | | Cite: | Binding site asymmetry in human transthyretin: insights from a joint neutron and X-ray crystallographic analysis using perdeuterated protein
IUCrJ, 1, 2014
|
|
3EIF
 
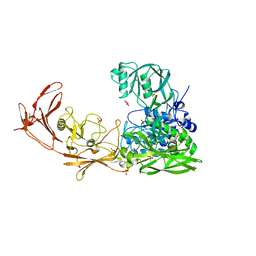 | | 1.9 angstrom crystal structure of the active form of the C5a peptidase from Streptococcus pyogenes (ScpA) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Cooney, J.C, Kagawa, T.F, O'Connell, M.R, Paoli, M, Mouat, P, O'Toole, P.W. | | Deposit date: | 2008-09-15 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Model for Substrate Interactions in C5a Peptidase from Streptococcus pyogenes: A 1.9 A Crystal Structure of the Active Form of ScpA
J.Mol.Biol., 386, 2009
|
|
3EC5
 
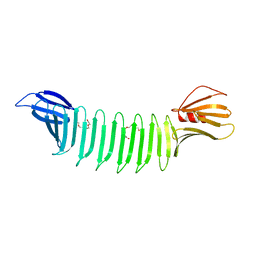 | | The crystal structure of Thioflavin-T (ThT) binding OspA mutant | | Descriptor: | Outer Surface Protein A, TETRAETHYLENE GLYCOL | | Authors: | Biancalana, M, Makabe, K, Koide, A, Koide, S. | | Deposit date: | 2008-08-28 | | Release date: | 2009-02-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanism of thioflavin-T binding to the surface of beta-rich peptide self-assemblies
J.Mol.Biol., 385, 2009
|
|
1LQ8
 
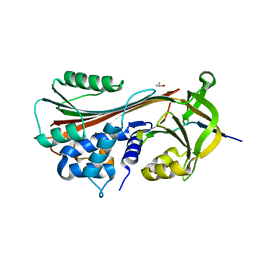 | | Crystal structure of cleaved protein C inhibitor | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huntington, J.A, Kjellberg, M, Stenflo, J. | | Deposit date: | 2002-05-09 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Protein C Inhibitor Provides Insights into Hormone Binding and Heparin Activation
Structure, 11, 2003
|
|
3EL0
 
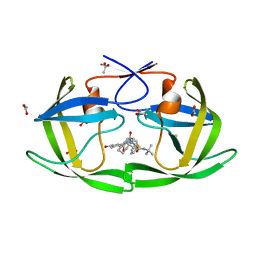 | |
4Q6P
 
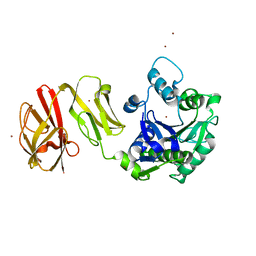 | | Structural analysis of the Zn-form I of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1LR5
 
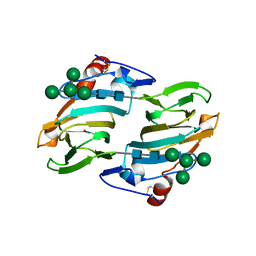 | | Crystal structure of auxin binding protein | | Descriptor: | Auxin binding protein 1, ZINC ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Woo, E.J, Marshall, J, Bauley, J, Chen, J.-G, Venis, M, Napier, R.M, Pickersgill, R.W. | | Deposit date: | 2002-05-14 | | Release date: | 2002-06-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of auxin-binding protein 1 in complex with auxin.
EMBO J., 21, 2002
|
|
4LAW
 
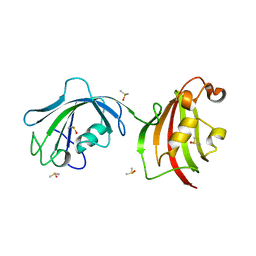 | | Crystal Structure Analysis of FKBP52, Crystal Form III | | Descriptor: | DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase FKBP4 | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
1LS6
 
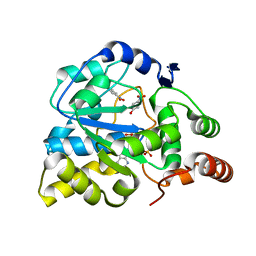 | | Human SULT1A1 complexed with PAP and p-Nitrophenol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, P-NITROPHENOL, aryl sulfotransferase | | Authors: | Gamage, N.U, Barnett, A.C, Tresillian, M, Latham, C.F, Liyou, N.E, McManus, M.E, Martin, J.L. | | Deposit date: | 2002-05-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a human carcinogen-converting enzyme, SULT1A1. Structural and kinetic implications of substrate inhibition.
J.Biol.Chem., 278, 2003
|
|
3AZC
 
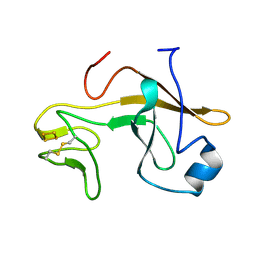 | | Crystal structure of the soluble part of cytochrome b6f complex iron-sulfur subunit from Thermosynechococcus elongatus BP-1 | | Descriptor: | Cytochrome b6-f complex iron-sulfur subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Veit, S, Takeda, K, Tsunoyama, Y, Roegner, M, Miki, K. | | Deposit date: | 2011-05-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a thermophilic cyanobacterial b(6)f-type Rieske protein
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1LT0
 
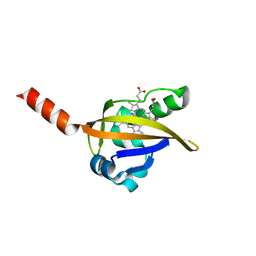 | | Crystal structure of the CN-bound BjFixL heme domain | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Sensor protein FixL | | Authors: | Hao, B, Isaza, C, Arndt, J, Soltis, M, Chan, M.K. | | Deposit date: | 2002-05-20 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based mechanism of O2 sensing and ligand discrimination by
the FixL heme domain of Bradyrhizobium japonicum
Biochemistry, 41, 2002
|
|
4Q88
 
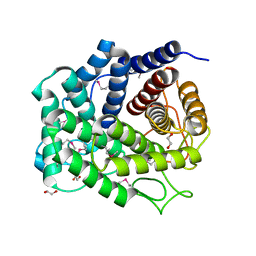 | | Glycosyl hydrolase family 88 from Bacteroides vulgatus | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Glycosyl hydrolase Family 88 from Bacteroides vulgatus
To be Published
|
|
4Q8V
 
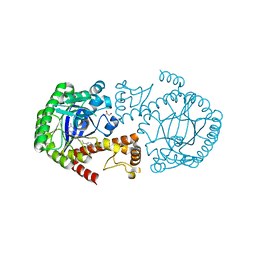 | | tRNA-Guanine Transglycosylase (TGT) in Complex with 4-[2-({6-Amino-8-oxo-1H,7H,8H-imidazo[4,5-g]quinazolin-2-yl}amino)ethyl]benzonitrile | | Descriptor: | 4-{2-[(6-amino-8-oxo-7,8-dihydro-1H-imidazo[4,5-g]quinazolin-2-yl)amino]ethyl}benzonitrile, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-04-28 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Addressing a New Subpocket of TGT by Elongated 2-Amino-lin-benzoguanines
To be Published
|
|
4LDV
 
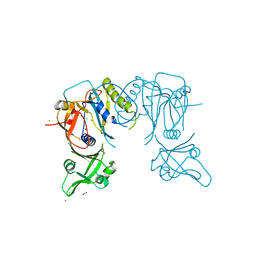 | | Crystal structure of the DNA binding domain of A. thailana auxin response factor 1 | | Descriptor: | Auxin response factor 1, CHLORIDE ION, FORMIC ACID, ... | | Authors: | boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
3EHQ
 
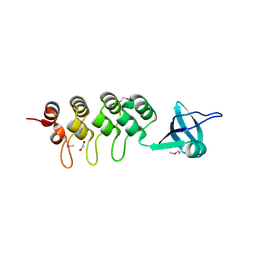 | | Crystal Structure of Human Osteoclast Stimulating Factor | | Descriptor: | 1,2-ETHANEDIOL, Osteoclast-stimulating factor 1 | | Authors: | Tong, S, Zhou, H, Gao, Y, Zhu, Z, Zhang, X, Teng, M, Niu, L. | | Deposit date: | 2008-09-14 | | Release date: | 2009-08-04 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of human osteoclast stimulating factor
Proteins, 75, 2009
|
|
3ZEP
 
 | | Crystal Structure of JAK3 Kinase Domain in Complex with a Pyrrolopyrazine-2-phenyl Ether Inhibitor | | Descriptor: | 2-[[(3R)-3-acetamido-2,3-dihydro-1H-inden-5-yl]oxy]-N-[(1S)-1-cyclopropylethyl]-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, GLYCEROL, TYROSINE-PROTEIN KINASE JAK3 | | Authors: | Kuglstatter, A, Jestel, A, Nagel, S, Boettcher, J, Blaesse, M. | | Deposit date: | 2012-12-06 | | Release date: | 2013-12-11 | | Last modified: | 2016-09-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of a Series of Novel 5H-Pyrrolo[2,3-B]Pyrazine-2-Phenyl Ethers, as Potent Jak3 Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4Q1K
 
 | |
3ROX
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Theophylline | | Descriptor: | Apolipoprotein A-I-binding protein, SULFATE ION, THEOPHYLLINE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
1LFZ
 
 | | OXY HEMOGLOBIN (25% METHANOL) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Biswal, B.K, Vijayan, M. | | Deposit date: | 2002-04-12 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of human oxy- and deoxyhaemoglobin at different levels of humidity: variability in the T state.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3RQ5
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis co-crystallized with ATP/Mg2+ and soaked with CoA | | Descriptor: | ADP/ATP-dependent NAD(P)H-hydrate dehydratase, COENZYME A, GLYCEROL | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
