1KR6
 
 | |
5U1Z
 
 | | X-ray structure of the WlarG aminotransferase, apo form, from Campylobacter jejune | | Descriptor: | CHLORIDE ION, Putative aminotransferase, SODIUM ION | | Authors: | Holden, H.M, Thoden, J.B, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
5RGM
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102142 (Mpro-x0708) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N'-acetyl-4,5,6,7-tetrahydro-1-benzothiophene-2-carbohydrazide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3BJZ
 
 | | Crystal structure of Pseudomonas aeruginosa phosphoheptose isomerase | | Descriptor: | CHLORIDE ION, Phosphoheptose isomerase, SULFATE ION | | Authors: | Walker, J.R, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-05 | | Release date: | 2007-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants.
J.Biol.Chem., 283, 2008
|
|
1KV4
 
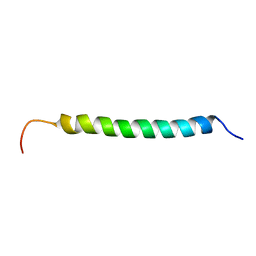 | | Solution structure of antibacterial peptide (Moricin) | | Descriptor: | moricin | | Authors: | Hemmi, H, Ishibashi, J, Hara, S, Yamakawa, M. | | Deposit date: | 2002-01-25 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of moricin, an antibacterial peptide, isolated from the silkworm Bombyx mori.
FEBS Lett., 518, 2002
|
|
5RGJ
 
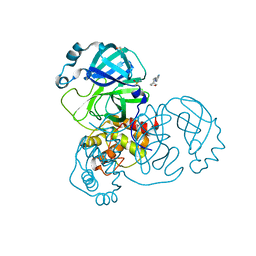 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1401276297 (Mpro-x0425) | | Descriptor: | (5S)-7-(pyrazin-2-yl)-2-oxa-7-azaspiro[4.4]nonane, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RGP
 
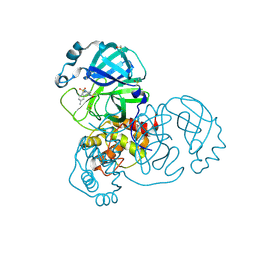 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102628 (Mpro-x0771) | | Descriptor: | 1-{4-[(2,4-dimethylphenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3BKM
 
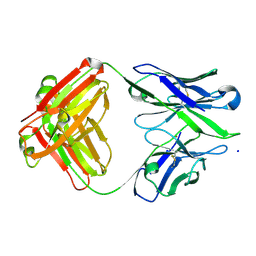 | | Structure of anti-amyloid-beta Fab WO2 (Form A, P212121) | | Descriptor: | SODIUM ION, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa, ... | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-07 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
1X59
 
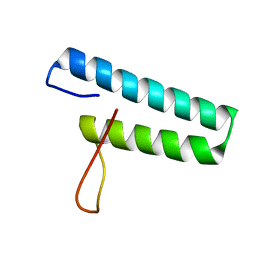 | | Solution structures of the WHEP-TRS domain of human histidyl-tRNA synthetase | | Descriptor: | Histidyl-tRNA synthetase | | Authors: | Nameki, N, Sasagawa, A, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the WHEP-TRS domain of human histidyl-tRNA synthetase
To be Published
|
|
5U3Z
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 10 | | Descriptor: | 6-[2-({cyclopropyl[4-(furan-3-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1KER
 
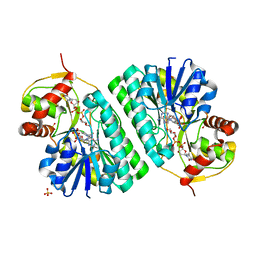 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-D-glucose bound | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
5UDV
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with iron | | Descriptor: | FE (III) ION, Lactate racemization operon protein LarE, PHOSPHATE ION, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U9W
 
 | | Structure of CNTnw N149L in the intermediate 3 state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-12-18 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.555 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
1WDL
 
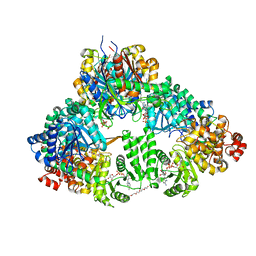 | | fatty acid beta-oxidation multienzyme complex from Pseudomonas fragi, form II (native4) | | Descriptor: | 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, 3-ketoacyl-CoA thiolase, ACETYL COENZYME *A, ... | | Authors: | Ishikawa, M, Tsuchiya, D, Oyama, T, Tsunaka, Y, Morikawa, K. | | Deposit date: | 2004-05-17 | | Release date: | 2004-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex
Embo J., 23, 2004
|
|
1LFG
 
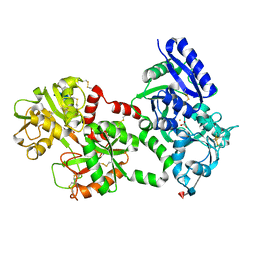 | | Structure of diferric human lactoferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Baker, E.N, Anderson, B.F, Haridas, M. | | Deposit date: | 1992-02-05 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human diferric lactoferrin refined at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
5UDQ
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, apo form | | Descriptor: | Lactate racemization operon protein LarE, PHOSPHATE ION, SULFATE ION | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TPU
 
 | | x-ray structure of the WlaRB TDP-quinovose 3,4-ketoisomerase from campylobacter jejuni | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Holden, H.M, Thoden, J.B, Li, J.Z, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinogradov, E, Schoenhofen, I.C, Gilbert, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
5UF3
 
 | |
7FAN
 
 | | Co-crystal Structure of Toxoplasma gondii Prolyl tRNA Synthetase (TgPRS) in complex with T35 and L-pro | | Descriptor: | 4-[(3S)-3-cyano-3-(1-methylcyclopropyl)-2-oxidanylidene-pyrrolidin-1-yl]-N-[[3-fluoranyl-5-(1-methylpyrazol-4-yl)phenyl]methyl]-6-methyl-pyridine-2-carboxamide, PROLINE, Prolyl-tRNA synthetase (ProRS) | | Authors: | Mishra, S, Malhotra, N, Yogavel, M, Sharma, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Targeting prolyl-tRNA synthetase via a series of ATP-mimetics to accelerate drug discovery against toxoplasmosis.
Plos Pathog., 19, 2023
|
|
2WBG
 
 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-oxa-(+)-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8R,8aR)-3-(octylimino)hexahydro[1,3]oxazolo[3,4-a]pyridine-5,6,7,8-tetrol, ACETATE ION, BETA-GLUCOSIDASE A | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-02-27 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
1LNT
 
 | | Crystal Structure of the Highly Conserved RNA Internal Loop of SRP | | Descriptor: | 5'-R(*CP*GP*GP*AP*AP*GP*CP*AP*GP*(CBV)P*GP*C)-3', 5'-R(*GP*CP*GP*UP*CP*AP*GP*GP*UP*CP*(CBV)P*G)-3', CALCIUM ION, ... | | Authors: | Deng, J, Xiong, Y, Pan, B, Sundaralingam, M. | | Deposit date: | 2002-05-03 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of an RNA dodecamer containing a fragment from SRP domain IV of Escherichia coli.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5TRU
 
 | | Structure of the first-in-class checkpoint inhibitor Ipilimumab bound to human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Ipilimumab Fab heavy chain, Ipilimumab Fab light chain | | Authors: | Ramagopal, U.A, Liu, W, Garrett-Thomson, S.C, Yan, Q, Srinivasan, M, Wong, S.C, Bell, A, Mankikar, S, Rangan, V.S, Deshpande, S, Bonanno, J.B, Korman, A.J, Almo, S.C. | | Deposit date: | 2016-10-27 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1WMF
 
 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (oxidized form, 1.73 angstrom) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
5TSO
 
 | |
7FAM
 
 | | Co-crystal Structure of Toxoplasma gondii Prolyl tRNA Synthetase (TgPRS) in complex with L97 and L-pro | | Descriptor: | 4-[(3S)-3-cyclopropyl-3-(hydroxymethyl)-2-oxidanylidene-pyrrolidin-1-yl]-N-[[3-fluoranyl-5-(1-methylpyrazol-4-yl)phenyl]methyl]-6-methyl-pyridine-2-carboxamide, PROLINE, Prolyl-tRNA synthetase (ProRS) | | Authors: | Mishra, S, Malhotra, N, Yogavel, M, Sharma, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Targeting prolyl-tRNA synthetase via a series of ATP-mimetics to accelerate drug discovery against toxoplasmosis.
Plos Pathog., 19, 2023
|
|
