7Y7E
 
 | | Structure of the Bacterial Ribosome with human tRNA Asp(ManQ34) and mRNA(GAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7G
 
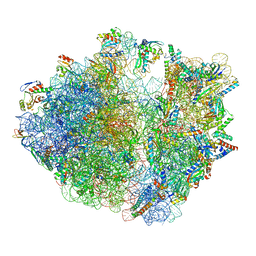 | | Structure of the Bacterial Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7H
 
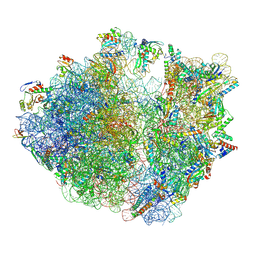 | | Structure of the Bacterial Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAC) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8AX1
 
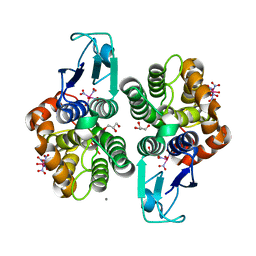 | |
1ZUJ
 
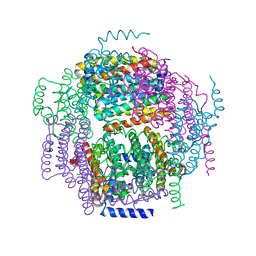 | | The crystal structure of the Lactococcus lactis MG1363 DpsA protein | | Descriptor: | hypothetical protein Llacc01001955 | | Authors: | Stillman, T.J, Upadhyay, M, Norte, V.A, Sedelnikova, S.E, Carradus, M, Tzokov, S, Bullough, P.A, Shearman, C.A, Gasson, M.J, Williams, C.H, Artymiuk, P.J, Green, J. | | Deposit date: | 2005-05-31 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding.
Mol.Microbiol., 57, 2005
|
|
1ZQ5
 
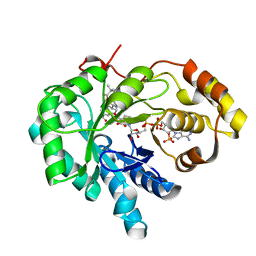 | | Crystal structure of human androgenic 17beta-hydroxysteroid dehydrogenase type 5 in complexed with a potent inhibitor EM1404 | | Descriptor: | 3-CARBOXAMIDO-1,3,5(10)-ESTRATRIEN-17(R)-SPIRO-2'(5',5'-DIMETHYL-6'OXO)TETRAHYDROPYRAN, ACETATE ION, Aldo-keto reductase family 1 member C3, ... | | Authors: | Qiu, W, Zhou, M, Ghanmi, D, Luu-The, V, Labrie, F, Lin, S.X. | | Deposit date: | 2005-05-18 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based inhibitor design for an enzyme that binds different steroids: a potent inhibitor for human type 5 17beta-hydroxysteroid dehydrogenase.
J.Biol.Chem., 282, 2007
|
|
1RCY
 
 | | RUSTICYANIN (RC) FROM THIOBACILLUS FERROOXIDANS | | Descriptor: | COPPER (II) ION, RUSTICYANIN | | Authors: | Walter, R.L, Friedman, A.M, Ealick, S.E, Blake II, R.C, Proctor, P, Shoham, M. | | Deposit date: | 1996-04-10 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple wavelength anomalous diffraction (MAD) crystal structure of rusticyanin: a highly oxidizing cupredoxin with extreme acid stability.
J.Mol.Biol., 263, 1996
|
|
6IO4
 
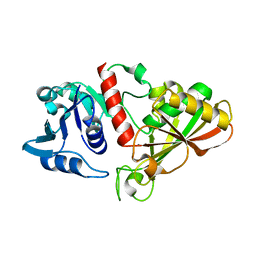 | |
6RX3
 
 | | Crystal structure of human syncytin 2 in post-fusion conformation | | Descriptor: | CHLORIDE ION, Syncytin-2 | | Authors: | Ruigrok, K, Backovic, M, Vaney, M.C, Rey, F.A. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Structures of the Post-fusion 6-Helix Bundle of the Human Syncytins and their Functional Implications.
J.Mol.Biol., 431, 2019
|
|
1BXP
 
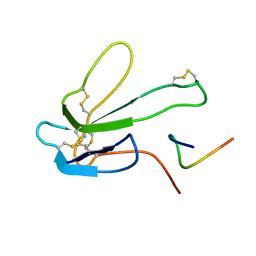 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF ALPHA-BUNGAROTOXIN WITH A LIBRARY DERIVED PEPTIDE, 20 STRUCTURES | | Descriptor: | ALPHA-BUNGAROTOXIN, PEPTIDE MET-ARG-TYR-TYR-GLU-SER-SER-LEU-LYS-SER-TYR-PRO-ASP | | Authors: | Scherf, T, Balass, M, Fuchs, S, Katchalski-Katzir, E, Anglister, J. | | Deposit date: | 1998-08-23 | | Release date: | 1999-01-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the complex of alpha-bungarotoxin with a library-derived peptide.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
6WZU
 
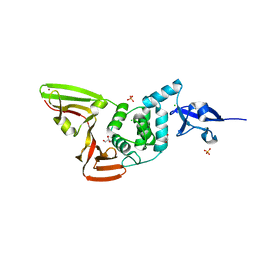 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , P3221 space group | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-14 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
6WPK
 
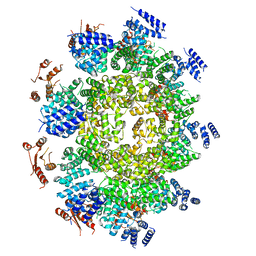 | |
5AVC
 
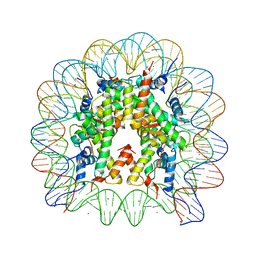 | | human nucleosome core particle | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Wakamori, M, Fujii, Y, Umehara, T, Yokoyama, S. | | Deposit date: | 2015-06-12 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation
Sci Rep, 5, 2015
|
|
8AIT
 
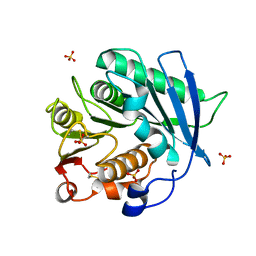 | | Crystal structure of cutinase PbauzCut from Pseudomonas bauzanensis | | Descriptor: | Cutinase, SULFATE ION | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
8AKN
 
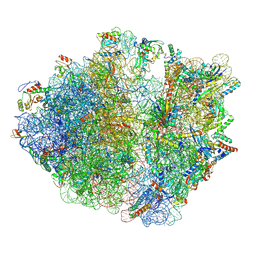 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the terminating ribosome | | Descriptor: | 16S ribosomal RNA, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-07-30 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
6QUC
 
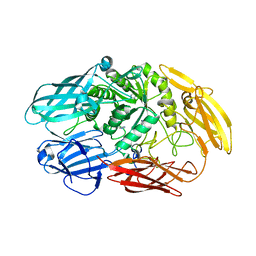 | | Truncated beta-galactosidase III from Bifidobacterium bifidum | | Descriptor: | Beta-galactosidase, CALCIUM ION, IMIDAZOLE | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
6GKE
 
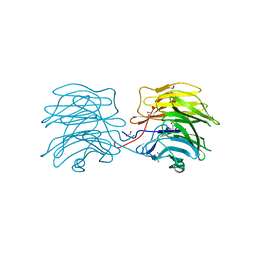 | | Aleuria aurantia lectin AAL N224Q mutant in complex with Fucalpha1-6GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, Fucose-specific lectin, GLYCEROL, ... | | Authors: | Houser, J, Wimmerova, M, Herrera, H, Mehta, A.S. | | Deposit date: | 2018-05-19 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural dynamics and applications of a fucose-binding lectin with enhanced binding to alpha-1,6-linked core fucosylation
To Be Published
|
|
1BLI
 
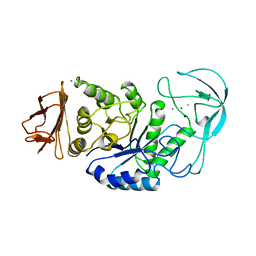 | | BACILLUS LICHENIFORMIS ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | Deposit date: | 1998-01-07 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activation of Bacillus licheniformis alpha-amylase through a disorder-->order transition of the substrate-binding site mediated by a calcium-sodium-calcium metal triad.
Structure, 6, 1998
|
|
6GM5
 
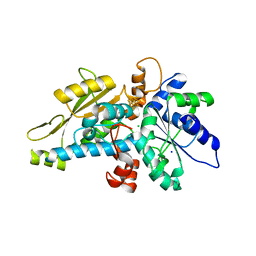 | | [FeFe]-hydrogenase HydA1 from Chlamydomonas reinhardtii,variant E141A | | Descriptor: | CHLORIDE ION, Fe-hydrogenase, IRON/SULFUR CLUSTER, ... | | Authors: | Duan, J, Engelbrecht, V, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases.
Nat Commun, 9, 2018
|
|
7TT3
 
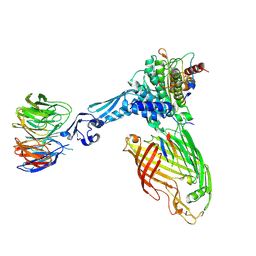 | | BamABCDE bound to substrate EspP class 5 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
1BT8
 
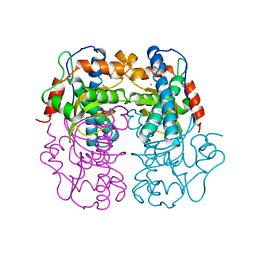 | | P.SHERMANII SOD(FE+3) PH 10.0 | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Schmidt, M. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Manipulating the coordination mumber of the ferric iron within the cambialistic superoxide dismutase of Propionibacterium shermanii by changing the pH-value A crystallographic analysis
Eur.J.Biochem., 262, 1999
|
|
7TT4
 
 | | BamABCDE bound to substrate EspP class 6 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
6QW2
 
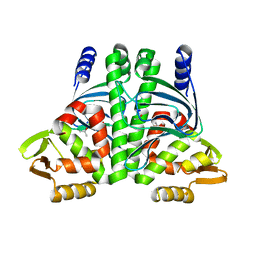 | | The Transcriptional Regulator PrfA-A218G mutant from Listeria Monocytogenes | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
6GLA
 
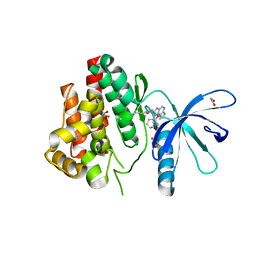 | | Crystal structure of JAK3 in complex with Compound 11 (FM481) | | Descriptor: | (~{E})-3-[5-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2(6),4,7,11-pentaen-4-yl)furan-2-yl]prop-2-enenitrile, 1,2-ETHANEDIOL, 1-phenylurea, ... | | Authors: | Chaikuad, A, Forster, M, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Development, Optimization, and Structure-Activity Relationships of Covalent-Reversible JAK3 Inhibitors Based on a Tricyclic Imidazo[5,4- d]pyrrolo[2,3- b]pyridine Scaffold.
J. Med. Chem., 61, 2018
|
|
7TSZ
 
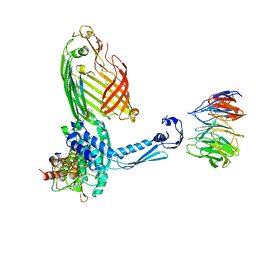 | | BamABCDE bound to substrate EspP class 1 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
