7H07
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0010744-001 | | Descriptor: | N-[(1S,2S)-1-(4-methoxyphenyl)-2-(1H-tetrazol-5-yl)propyl]-7H-pyrrolo[2,3-d]pyrimidine-4-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H0F
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011207-001 | | Descriptor: | N-cyclopropyl-4-{[(1R)-1-(1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~-benzothiopyran-7-yl)-2-methylpropyl]amino}-7H-pyrrolo[2,3-d]pyrimidine-5-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H0Y
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013258-001 | | Descriptor: | 4-{[(1S)-1-(1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~-benzothiopyran-7-yl)-2-methylpropyl]amino}-N-(1-methylazetidin-3-yl)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
2BOI
 
 | | 1.1A Structure of Chromobacterium Violaceum Lectin CV2L in Complex with alpha-methyl-fucoside | | Descriptor: | CALCIUM ION, CV-IIL LECTIN, methyl alpha-L-fucopyranoside | | Authors: | Pokorna, M, Cioci, G, Perret, S, Rebuffet, E, Adam, J, Gilboa-Garber, N, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2005-04-12 | | Release date: | 2006-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Unusual Entropy Driven Affinity of Chromobacter Violaceum Lectin Cv-Iil Towards Fucose and Mannose
Biochemistry, 45, 2006
|
|
1GWO
 
 | |
6UTT
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with calcium | | Descriptor: | ATP-dependent sacrificial sulfur transferase LarE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Fellner, M, Huizenga, K, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystallographic characterization of a tri-Asp metal-binding site at the three-fold symmetry axis of LarE.
Sci Rep, 10, 2020
|
|
1GTK
 
 | | Time-resolved and static-ensemble structural chemistry of hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, PORPHOBILINOGEN DEAMINASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | Deposit date: | 2002-01-16 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Time-Resolved and Static-Ensemble Structural Chemistry of Hydroxymethylbilane Synthase
Faraday Discuss., 122, 2003
|
|
1GX2
 
 | |
4BGH
 
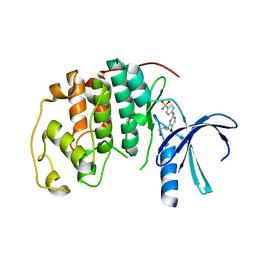 | | Crystal Structure of CDK2 in complex with pan-CDK Inhibitor | | Descriptor: | 4-((5-BROMO-4-(PROP-2-YN-1-YLAMINO)PYRIMIDIN-2-YL)AMINO)BENZENESULFONAMIDE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Luecking, U, Jautelat, R, Krueger, M, Brumby, T, Lienau, P, Schaefer, M, Briem, H, Schulze, J, Hillisch, A, Reichel, A, Siemeister, G. | | Deposit date: | 2013-03-26 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Lab Oddity Prevails: Discovery of Pan-Cdk Inhibitor (R)- S-Cyclopropyl-S-(4-{[4-{[(1R,2R)-2-Hydroxy-1-Methylpropyl]Oxy}-5-(Trifluoromethyl)Pyrimidin-2-Yl]Amino}Phenyl)Sulfoximide (Bay 1000394) for the Treatment of Cancer.
Chemmedchem, 8, 2013
|
|
7AK3
 
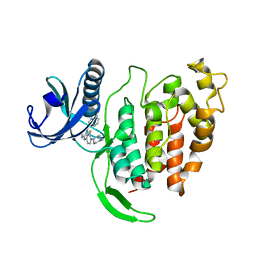 | | CLK1 bound with CAF052 | | Descriptor: | Dual specificity protein kinase CLK1, ~{N}-[3-fluoranyl-4-(4-methylpiperazin-1-yl)phenyl]-4-pyrazolo[1,5-b]pyridazin-3-yl-pyrimidin-2-amine | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-29 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Inhibitor Identifications Reveal Targeting Opportunity for the Atypical MAPK Kinase ERK3.
Int J Mol Sci, 21, 2020
|
|
7GZC
 
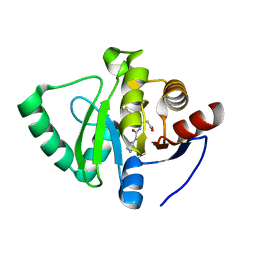 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000588-001 | | Descriptor: | (3R)-3-(4-fluorophenyl)-3-[(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.185 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
8CTH
 
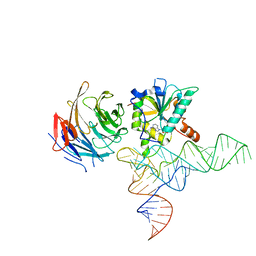 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Phe) complex | | Descriptor: | Phe-tRNA, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(7)-)-methyltransferase, ... | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fishcer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
7GZO
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0008338-001 | | Descriptor: | 3-cyclopropyl-N-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-L-alanine, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
6V08
 
 | | Crystal structure of human recombinant Beta-2 glycoprotein I (hrB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
7GZX
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011176-001 | | Descriptor: | 7-[(1S)-2-methyl-1-{[(6M)-6-{5-[(methylamino)methyl]furan-3-yl}-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}propyl]-3,4-dihydro-1lambda~6~-benzothiopyran-1,1(2H)-dione, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H0H
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011204-001 | | Descriptor: | N-ethyl-4-{[(1S)-2-methyl-1-(3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)propyl]amino}-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
3Q3C
 
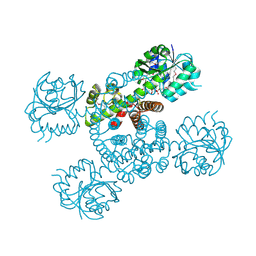 | | Crystal structure of a serine dehydrogenase from Pseudomonas aeruginosa pao1 in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable 3-hydroxyisobutyrate dehydrogenase | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
8CTI
 
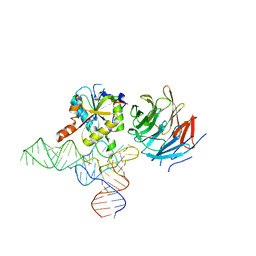 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Val) complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4, tRNA-Val-TAC-2-1 | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fischer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
7H14
 
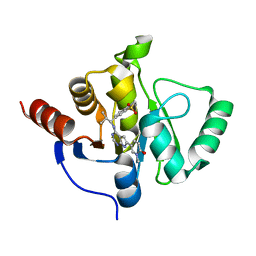 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013383-001 | | Descriptor: | 7-[(1R)-2-methyl-1-{[(6M)-6-(1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}propyl]-3,4-dihydro-1lambda~6~-benzothiopyran-1,1(2H)-dione, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H1F
 
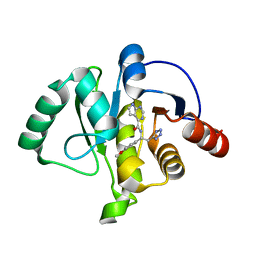 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0015381-001 | | Descriptor: | N-[(1R)-1-(4-methoxyphenyl)-2-(1H-tetrazol-5-yl)ethyl]-7H-pyrrolo[2,3-d]pyrimidine-4-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
5NXI
 
 | | Carbonic Anhydrase II Inhibitor RA2 | | Descriptor: | 4-[(4-bromophenyl)sulfonylamino]benzenesulfonamide, CITRIC ACID, Carbonic anhydrase 2, ... | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXO
 
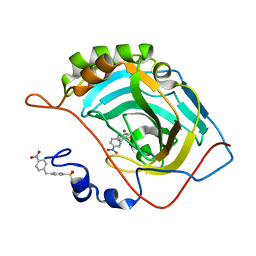 | | Carbonic Anhydrase II Inhibitor RA6 | | Descriptor: | 4-[(4-nitrophenyl)methyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXV
 
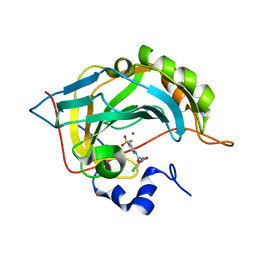 | | Carbonic Anhydrase II Inhibitor RA8 | | Descriptor: | 2-(4-chloranyl-5-methyl-3-nitro-pyrazol-1-yl)-~{N}-(4-sulfamoylphenyl)ethanamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NY3
 
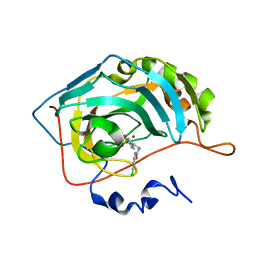 | | Carbonic Anhydrase II Inhibitor RA11 | | Descriptor: | 1-(4-chlorophenyl)-3-[2-(4-sulfamoylphenyl)ethyl]urea, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
6IZK
 
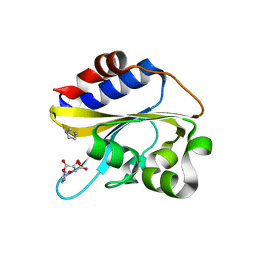 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, IMIDAZOLE, L(+)-TARTARIC ACID, ... | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2018-12-19 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus
To Be Published
|
|
