4HUK
 
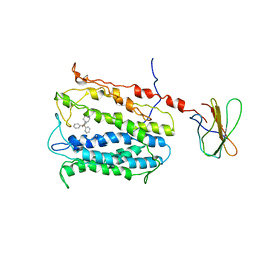 | | MATE transporter NorM-NG in complex with TPP and monobody | | Descriptor: | Multidrug efflux protein, Protein B, TETRAPHENYLPHOSPHONIUM | | Authors: | Lu, M. | | Deposit date: | 2012-11-02 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structures of a Na+-coupled, substrate-bound MATE multidrug transporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HHT
 
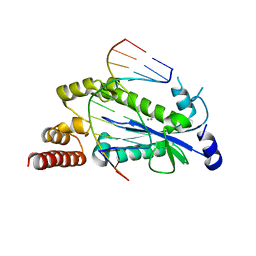 | |
1Z14
 
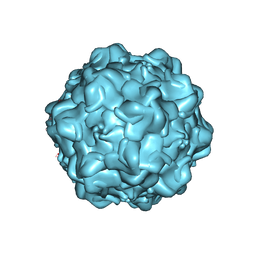 | | Structural Determinants of Tissue Tropism and In Vivo Pathogenicity for the Parvovirus Minute Virus of Mice | | Descriptor: | VP2 | | Authors: | Kontou, M, Govindasamy, L, Nam, H.J, Bryant, N, Llamas-Saiz, A.L, Foces-Foces, C, Hernando, E, Rubio, M.P, McKenna, R, Almendral, J.M, Agbandje-McKenna, M. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice.
J.Virol., 79, 2005
|
|
7PGA
 
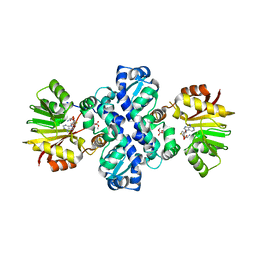 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
1PHR
 
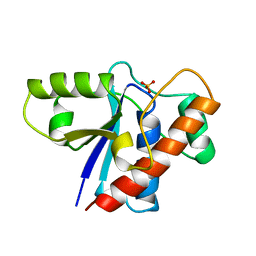 | | THE CRYSTAL STRUCTURE OF A LOW MOLECULAR PHOSPHOTYROSINE PROTEIN PHOSPHATASE | | Descriptor: | LOW MOLECULAR WEIGHT PHOSPHOTYROSINE PROTEIN PHOSPHATASE, SULFATE ION | | Authors: | Su, X.-D, Taddei, N, Stefani, M, Ramponi, G, Nordlund, P. | | Deposit date: | 1994-07-05 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a low-molecular-weight phosphotyrosine protein phosphatase.
Nature, 370, 1994
|
|
4MDQ
 
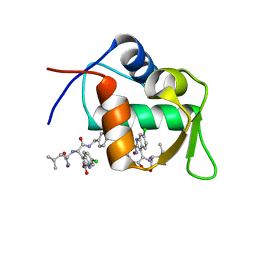 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-[(1R)-2-(benzylamino)-1-{[(2S)-1-(hydroxyamino)-4-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl]-6-chloro-N-hydroxy-1H-indole-2-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Bista, M, Popowicz, G, Holak, T.A. | | Deposit date: | 2013-08-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
8CLR
 
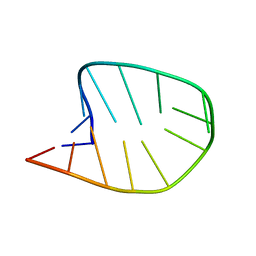 | | Integrated NMR/MD structure determination of a dynamic and thermodynamically stable CUUG RNA tetraloop | | Descriptor: | RNA hairpin with CUUG tetraloop | | Authors: | Oxenfarth, A, Kuemmerer, F, Bottaro, S, Schnieders, R, Pinter, G, Jonker, H.R.A, Fuertig, B, Richter, C, Blackledge, M, Lindorff-Larsen, K, Schwalbe, H. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Integrated NMR/Molecular Dynamics Determination of the Ensemble Conformation of a Thermodynamically Stable CUUG RNA Tetraloop.
J.Am.Chem.Soc., 145, 2023
|
|
6TY4
 
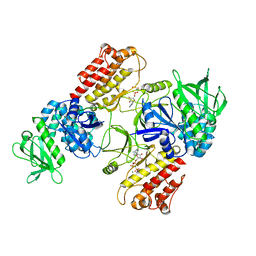 | | FAK structure with AMP-PNP from single particle analysis of 2D crystals | | Descriptor: | Focal adhesion kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Acebron, I, Righetto, R, Biyani, N, Chami, M, Boskovic, J, Stahlberg, H, Lietha, D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.96 Å) | | Cite: | Structural basis of Focal Adhesion Kinase activation on lipid membranes.
Embo J., 39, 2020
|
|
3WSV
 
 | |
7F02
 
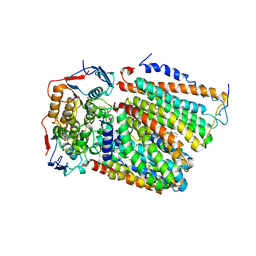 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
5MHM
 
 | |
3WF8
 
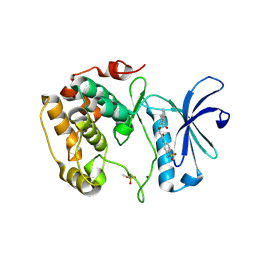 | | Crystal structure of S6K1 kinase domain in complex with a quinoline derivative 2-oxo-2-[(4-sulfamoylphenyl)amino]ethyl 7,8,9,10-tetrahydro-6H-cyclohepta[b]quinoline-11-carboxylate | | Descriptor: | 2-oxo-2-[(4-sulfamoylphenyl)amino]ethyl 7,8,9,10-tetrahydro-6H-cyclohepta[b]quinoline-11-carboxylate, GLYCEROL, Ribosomal protein S6 kinase beta-1, ... | | Authors: | Niwa, H, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
3WTZ
 
 | |
7F04
 
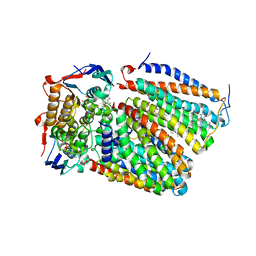 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with Heme and ATP. | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
3WU3
 
 | | Reduced-form structure of E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
7F03
 
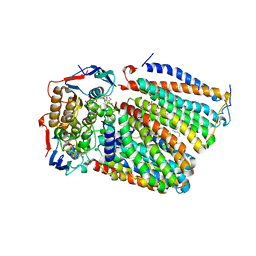 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
3WG8
 
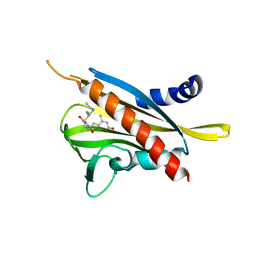 | | Crystal structure of the abscisic acid receptor PYR1 in complex with an antagonist AS6 | | Descriptor: | (2Z,4E)-5-[(1S)-3-(hexylsulfanyl)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Akiyama, T, Sue, M, Takeuchi, J, Okamoto, M, Muto, T, Endo, A, Nambara, E, Hirai, N, Ohnishi, T, Cutler, S.R, Todoroki, Y, Yajima, S. | | Deposit date: | 2013-07-31 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Designed abscisic acid analogs as antagonists of PYL-PP2C receptor interactions
Nat.Chem.Biol., 10, 2014
|
|
1PYS
 
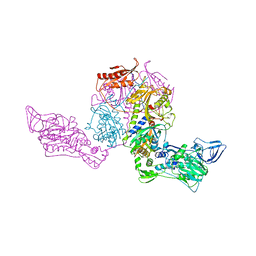 | | PHENYLALANYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | Descriptor: | MAGNESIUM ION, PHENYLALANYL-TRNA SYNTHETASE | | Authors: | Safro, M, Mosyak, L, Goldgur, Y, Reshetnikova, L, Delarue, M. | | Deposit date: | 1996-11-14 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of phenylalanyl-tRNA synthetase from Thermus thermophilus.
Nat.Struct.Biol., 2, 1995
|
|
6HAH
 
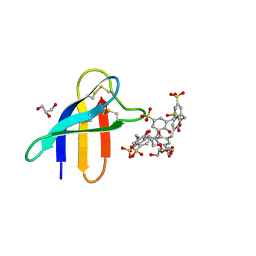 | | Crystal structure of PAF - p-sulfonatocalix[6]arene complex | | Descriptor: | 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, GLYCEROL, Pc24g00380 protein, ... | | Authors: | Alex, J.M, Rennie, M, Engilberge, S, Batta, G, Crowley, P.B. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Calixarene-mediated assembly of a small antifungal protein.
Iucrj, 6, 2019
|
|
6ZFH
 
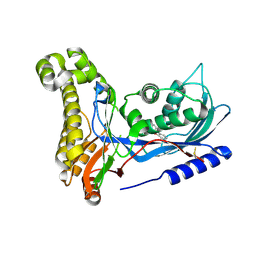 | | Structure of human galactokinase in complex with galactose and 2'-(benzo[d]oxazol-2-ylamino)-7',8'-dihydro-1'H-spiro[cyclopentane-1,4'-quinazolin]-5'(6'H)-one | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclopentane]-5-one, Galactokinase, beta-D-galactopyranose | | Authors: | Bezerra, G.A, Mackinnon, S, Zhang, M, Foster, W, Bailey, H, Arrowsmith, C, Edwards, A, Bountra, C, Lai, K, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.439 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
5MLK
 
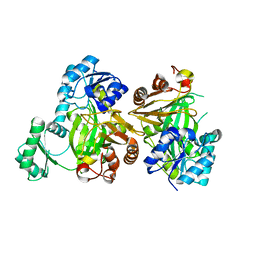 | |
7PHD
 
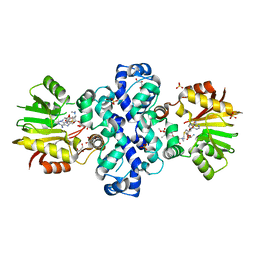 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with a region from 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein, GLYCEROL, S-ADENOSYLMETHIONINE, ... | | Authors: | Grocholski, T, Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
6U3T
 
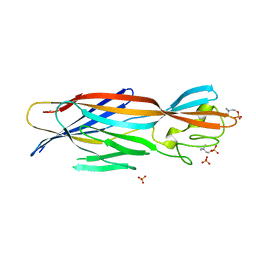 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
7PHE
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
6U4P
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-26 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
