5R2A
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry H02, DMSO-free | | Descriptor: | 3-cyclopentyl-1-(piperazin-1-yl)propan-1-one, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.049 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
2G55
 
 | | Anomalous substructure of trypsin (P3121) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cationic trypsin | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5R2N
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 11, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.078 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R33
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 27, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3I
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 42, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RC7
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library F11a | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6TLP
 
 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, CITRATE ANION, Casein kinase II subunit alpha, ... | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
5RCN
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 08 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RD3
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 25 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RDK
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 44 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6T9Z
 
 | | Nidocarborane inhibitor of Carbonic Anhydrase IX | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, Nidocarborane, ... | | Authors: | Brynda, J, Rezacova, P, Kugler, M, Gruner, B. | | Deposit date: | 2019-10-29 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Sulfonamido carboranes as highly selective inhibitors of cancer-specific carbonic anhydrase IX.
Eur.J.Med.Chem., 200, 2020
|
|
5RE0
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 57 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6GRE
 
 | |
5NC2
 
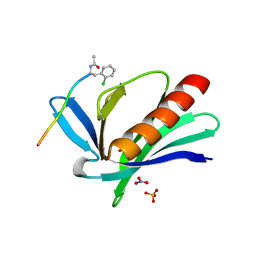 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPPTEDEL-NH2 | | Descriptor: | Ac-[2-Cl-F]PPPPTEDEL-NH2, NITRATE ION, Protein enabled homolog, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-03-03 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2G4M
 
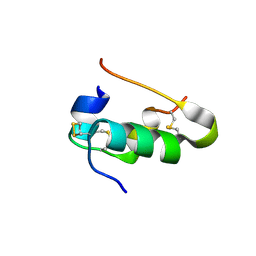 | | Insulin collected at 2.0 A wavelength | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2G5M
 
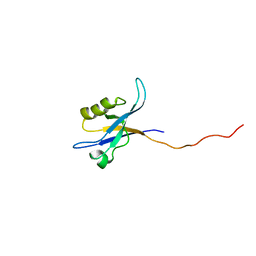 | | Spinophilin PDZ domain | | Descriptor: | Neurabin-2 | | Authors: | Kelker, M.S, Peti, W. | | Deposit date: | 2006-02-23 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for spinophilin-neurabin receptor interaction.
Biochemistry, 46, 2007
|
|
6ZTV
 
 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | 1,2-ETHANEDIOL, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
8PD6
 
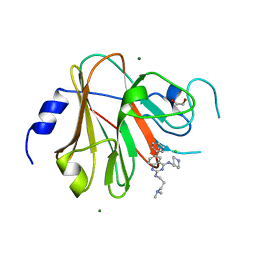 | | Crystal structure of the TRIM58 PRY-SPRY domain in complex with TRIM-473 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase TRIM58, ... | | Authors: | Renatus, M, Hoegenauer, K, Schroeder, M. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Ligands for TRIM58, a Novel Tissue-Selective E3 Ligase.
Acs Med.Chem.Lett., 14, 2023
|
|
8PD4
 
 | |
8PHE
 
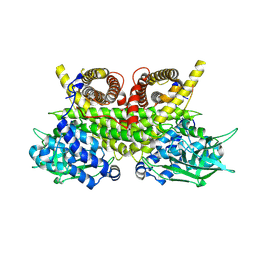 | | ACAD9-WT in complex with ECSIT-CTER | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, Evolutionarily conserved signaling intermediate in Toll pathway | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
5D7A
 
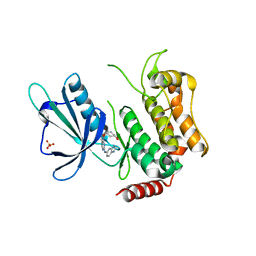 | | Crystal structure of the kinase domain of TRAF2 and NCK-interacting protein kinase with NCB-0846 | | Descriptor: | SULFATE ION, TRAF2 and NCK-interacting protein kinase, cis-4-{[2-(1H-benzimidazol-5-ylamino)quinazolin-8-yl]oxy}cyclohexanol | | Authors: | Ohbayashi, N, Kukimoto-Niino, M, Yamada, T, Shirouzu, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | TNIK inhibition abrogates colorectal cancer stemness
Nat Commun, 7, 2016
|
|
8PHF
 
 | | Cryo-EM structure of human ACAD9-S191A | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
5LNZ
 
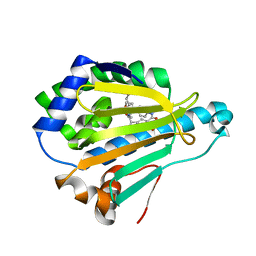 | | HSP90 WITH indazole derivative | | Descriptor: | Heat shock protein HSP 90-alpha, ~{N}3-butyl-~{N}3,~{N}5-dimethyl-~{N}5-(4-morpholin-4-ylphenyl)-6-oxidanyl-2~{H}-indazole-3,5-dicarboxamide | | Authors: | Graedler, U, Amaral, M, Schuetz, D. | | Deposit date: | 2016-08-08 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Ligand Desolvation Steers On-Rate and Impacts Drug Residence Time of Heat Shock Protein 90 (Hsp90) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6W0P
 
 | | Putative kojibiose phosphorylase from human microbiome | | Descriptor: | Kojibiose phosphorylase | | Authors: | Dementiev, A, Osipiuk, J, Endres, M, Wakatsuki, S, Hess, M, Joachimiak, A. | | Deposit date: | 2020-03-02 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Putative kojibiose phosphorylase from human microbiome
to be published
|
|
6YQ1
 
 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH N-Methyl-N-(3-{[2-(2-oxo-1,2,3,4-tetrahydro-quinolin-6-ylamino)-5-trifluoromethyl-pyrimidin-4-ylamino]-methyl}-pyridin-2-yl)-methanesulfonamide | | Descriptor: | Focal adhesion kinase 1, SODIUM ION, SULFATE ION, ... | | Authors: | Musil, D, Heinrich, T, Amaral, M. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Structure-kinetic relationship reveals the mechanism of selectivity of FAK inhibitors over PYK2.
Cell Chem Biol, 28, 2021
|
|
