3R7T
 
 | | Crystal Structure of Adenylosuccinate Synthetase from Campylobacter jejuni | | Descriptor: | Adenylosuccinate synthetase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Kim, Y, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-23 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Adenylosuccinate Synthetase from
Campylobacter jejuni
To be Published
|
|
6HE2
 
 | | Crystal structure of an open conformation of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in complex with 2-HIB-AMP and CoA | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, ADENOSINE MONOPHOSPHATE, COENZYME A, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
3RE8
 
 | |
3RGF
 
 | | Crystal Structure of human CDK8/CycC | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Blaesse, M, Huber, R, Maskos, K. | | Deposit date: | 2011-04-08 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of CDK8/CycC Implicates Specificity in the CDK/Cyclin Family and Reveals Interaction with a Deep Pocket Binder.
J.Mol.Biol., 412, 2011
|
|
3RI8
 
 | | Xylanase C from Aspergillus kawachii D37N mutant | | Descriptor: | Endo-1,4-beta-xylanase 3 | | Authors: | Fushinobu, S, Uno, T, Kitaoka, M, Hayashi, K, Matsuzawa, H, Wakagi, T. | | Deposit date: | 2011-04-13 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational analysis of fungal family 11 xylanases on pH optimum determination
J.APPL.GLYOSCI., 58, 2011
|
|
6K0N
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0P
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1045A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
3RKV
 
 | | C-terminal domain of protein C56C10.10, a putative peptidylprolyl isomerase, from Caenorhabditis elegans | | Descriptor: | putative peptidylprolyl isomerase | | Authors: | Osipiuk, J, Tesar, C, Gu, M, Van Oosten-Hawle, P, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | C-terminal domain of protein C56C10.10, a putative peptidylprolyl isomerase, from Caenorhabditis elegans
To be Published
|
|
6K2S
 
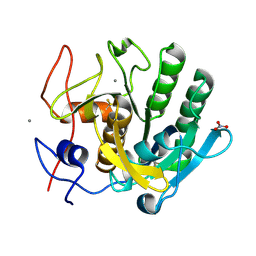 | |
3RRF
 
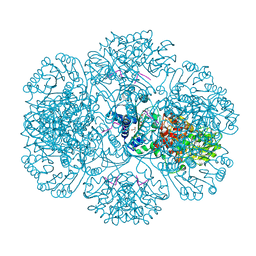 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, GLYCEROL, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-04-29 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTD
 
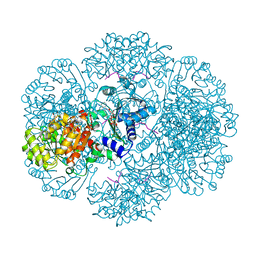 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADH and ADP. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE-5'-DIPHOSPHATE, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
7OFU
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 3, 4-Dihydroxybenzoic acid, methyl ester | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Ewert, W, Werner, N, Falke, S, Guenther, S, Reinke, P, Sprenger, J, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Wolf, M, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
6KA9
 
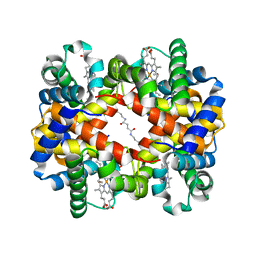 | | Crosslinked alpha(Fe-CO)-beta(Ni) human hemoglobin A in the T quaternary structure at 95 K: Dark | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-21 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7OFS
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 4-(2-hydroxyethyl)phenol | | Descriptor: | 4-(2-hydroxyethyl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Ewert, W, Sprenger, J, Koua, F, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Wolf, M, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
6KA1
 
 | | E.coli Malate dehydrogenase | | Descriptor: | Malate dehydrogenase | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | MDH is a major silver target in E. coli
To Be Published
|
|
7OFT
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with p-hydroxybenzaldehyde | | Descriptor: | CHLORIDE ION, P-HYDROXYBENZALDEHYDE, POTASSIUM ION, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Ewert, W, Sprenger, J, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Wolf, M, Yefanov, O, Gelisio, L, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
3R55
 
 | | Crystal structure of the complex of goat lactoperoxidase with Pyrazinamide at 2.1 A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, R.P, Pandey, N, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-03-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with Pyrazinamide at 2.1 A resolution
To be Published
|
|
3RQ2
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis co-crystallized with ATP/Mg2+ and soaked with NADH | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3R69
 
 | | Molecular analysis of the interaction of the HDL-receptor SR-BI with the PDZ3 domain of its adaptor protein PDZK1 | | Descriptor: | CITRIC ACID, Na(+)/H(+) exchange regulatory cofactor NHE-RF3, Scavenger receptor class B member 1 | | Authors: | Kocher, O, Birrane, G, Krieger, M. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Identification of the PDZ3 Domain of the Adaptor Protein PDZK1 as a Second, Physiologically Functional Binding Site for the C Terminus of the High Density Lipoprotein Receptor Scavenger Receptor Class B Type I.
J.Biol.Chem., 286, 2011
|
|
6KEZ
 
 | | Crystal structure of GAPDH/CP12/PRK complex from Arabidopsis thaliana | | Descriptor: | Calvin cycle protein CP12-2, Glyceraldehyde-3-phosphate dehydrogenase GAPA1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yu, A, Xie, Y, Li, M. | | Deposit date: | 2019-07-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Photosynthetic Phosphoribulokinase Structures: Enzymatic Mechanisms and the Redox Regulation of the Calvin-Benson-Bassham Cycle.
Plant Cell, 32, 2020
|
|
3RRE
 
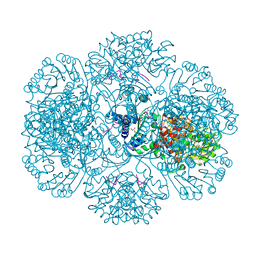 | | Crystal Structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, GLYCEROL, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-04-29 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
6KEW
 
 | |
6KFG
 
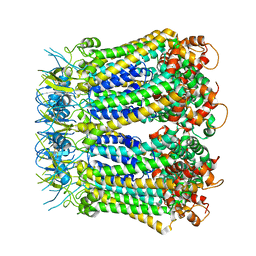 | | Undocked INX-6 hemichannel in detergent | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
6KG7
 
 | | Cryo-EM Structure of the Mammalian Tactile Channel Piezo2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Piezo-type mechanosensitive ion channel component 2 | | Authors: | Wang, L, Zhou, H, Zhang, M, Liu, W, Deng, T, Zhao, Q, Li, Y, Lei, J, Li, X, Xiao, B. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and mechanogating of the mammalian tactile channel PIEZO2.
Nature, 573, 2019
|
|
6JY6
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
