3C03
 
 | | Crystal structure of the EscU C-terminal domain with P263A mutation,space group P 1 21 1 | | Descriptor: | EscU, PROLINE | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
6K64
 
 | | Application of anti-helix antibodies in protein structure determination (8188-3LRH) | | Descriptor: | 3LRH intrabody, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K6A
 
 | | Application of anti-helix antibodies in protein structure determination (8188cys-3LRHcys) | | Descriptor: | 3LRH intrabody, Engineered Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-02 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3BZL
 
 | | Crystal structural of native EscU C-terminal domain | | Descriptor: | EscU, FORMIC ACID, SODIUM ION | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
7ZBO
 
 | | Amine Dehydrogenase MATOUAmDH2 in complex with NADP+ | | Descriptor: | Amine Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bennett, M, Ducrot, L, Vergne-Vaxelaire, C, Grogan, G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Mutation of the Native Amine Dehydrogenase MATOUAmDH2.
Chembiochem, 23, 2022
|
|
3BZX
 
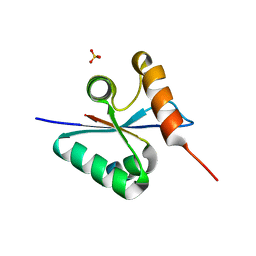 | | Crystal structure of the mutated H265A EscU C-terminal domain | | Descriptor: | EscU, SULFATE ION | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
7EPO
 
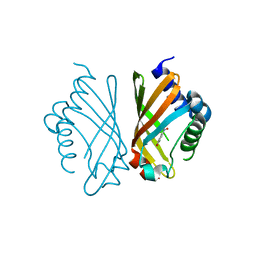 | | Ketosteroid Isomerase KSI with 5-nitrobenzoxazole (5NBI) | | Descriptor: | 5-nitro-1,2-benzoxazole, SnoaL-like domain-containing protein | | Authors: | Liang, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-04-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization and Kemp eliminase activity of the Mycobacterium smegmatis Ketosteroid Isomerase.
Biochem.Biophys.Res.Commun., 560, 2021
|
|
3C0T
 
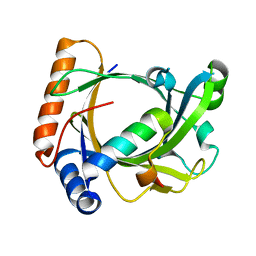 | | Structure of the Schizosaccharomyces pombe Mediator subcomplex Med8C/18 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 18, Mediator of RNA polymerase II transcription subunit 8 | | Authors: | Lariviere, L, Seizl, M, van Wageningen, S, Roether, S, van de Pasch, L, Feldmann, H, Straesser, K, Hahn, S, Holstege, C.P, Cramer, P. | | Deposit date: | 2008-01-21 | | Release date: | 2008-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-system correlation identifies a gene regulatory Mediator submodule
Genes Dev., 22, 2008
|
|
7EPN
 
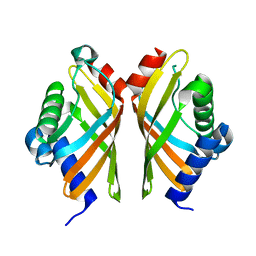 | | Ketosteroid Isomerase KSI native | | Descriptor: | SnoaL-like domain-containing protein | | Authors: | Liang, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-04-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization and Kemp eliminase activity of the Mycobacterium smegmatis Ketosteroid Isomerase.
Biochem.Biophys.Res.Commun., 560, 2021
|
|
7Z51
 
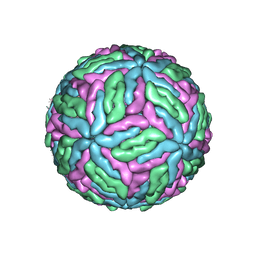 | | Tick-borne encephalitis virus Kuutsalo-14 | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Pulkkinen, L.I.A, Barrass, S.V, Anastasina, M, Butcher, S.J. | | Deposit date: | 2022-03-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular Organisation of Tick-Borne Encephalitis Virus.
Viruses, 14, 2022
|
|
6KF3
 
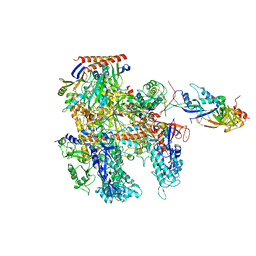 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
3C2Q
 
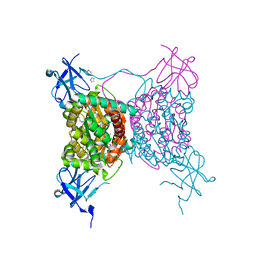 | | Crystal structure of conserved putative LOR/SDH protein from Methanococcus maripaludis S2 | | Descriptor: | IMIDAZOLE, NICKEL (II) ION, Uncharacterized conserved protein | | Authors: | Duke, N, Gu, M, Mulligan, R, Conrad, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-25 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of conserved putative LOR/SDH protein from Methanococcus maripaludis S2
To be Published
|
|
7F8X
 
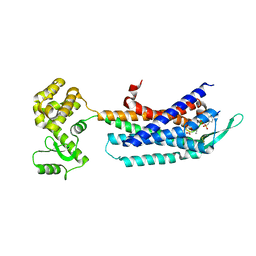 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with NN9056 | | Descriptor: | ASP-SMF-NLE-GLY-TRP-NLE-OEM-MEA-NH2 (NN9056), Cholecystokinin receptor type A,Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7Z6H
 
 | | Structure of DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1,Cell cycle checkpoint protein RAD17, Checkpoint protein HUS1, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-05-04 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction.
Nucleic Acids Res., 50, 2022
|
|
3BS8
 
 | | Crystal structure of Glutamate 1-Semialdehyde Aminotransferase complexed with pyridoxamine-5'-phosphate From Bacillus subtilis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Ge, H, Fan, J, Teng, M, Niu, L. | | Deposit date: | 2007-12-22 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Glutamate1-semialdehyde aminotransferase from Bacillus subtilis with bound pyridoxamine-5'-phosphate
Biochem.Biophys.Res.Commun., 402, 2010
|
|
3BSU
 
 | | Hybrid-binding domain of human RNase H1 in complex with 12-mer RNA/DNA | | Descriptor: | DNA (5'-D(*DGP*DAP*DAP*DTP*DCP*DAP*DGP*DGP*(5IU)P*DGP*DTP*DC)-3'), MAGNESIUM ION, RNA (5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3'), ... | | Authors: | Nowotny, M, Cerritelli, S.M, Ghirlando, R, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2007-12-26 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD.
Embo J., 27, 2008
|
|
3BTT
 
 | | THE CRYSTAL STRUCTURES OF THE COMPLEXES BETWEEN BOVINE BETA-TRYPSIN AND TEN P1 VARIANTS OF BPTI | | Descriptor: | CALCIUM ION, PROTEIN (PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN), ... | | Authors: | Helland, R, Otlewski, J, Sundheim, O, Dadlez, M, Smalas, A.O. | | Deposit date: | 1999-03-10 | | Release date: | 2000-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of the complexes between bovine beta-trypsin and ten P1 variants of BPTI.
J.Mol.Biol., 287, 1999
|
|
3C9E
 
 | | Crystal structure of the cathepsin K : chondroitin sulfate complex. | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Cathepsin K, ... | | Authors: | Kienetz, M, Cherney, M.M, James, M.N.G, Bromme, D. | | Deposit date: | 2008-02-15 | | Release date: | 2008-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal and molecular structures of a cathepsin K:chondroitin sulfate complex.
J.Mol.Biol., 383, 2008
|
|
7YWX
 
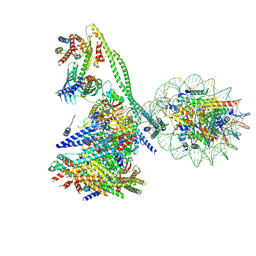 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7ER9
 
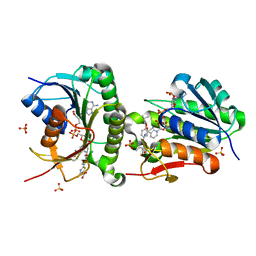 | | Crystal structure of human Biliverdin IX-beta reductase B with Febuxostat (TEI) | | Descriptor: | 2-(3-CYANO-4-ISOBUTOXY-PHENYL)-4-METHYL-5-THIAZOLE-CARBOXYLIC ACID, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ERE
 
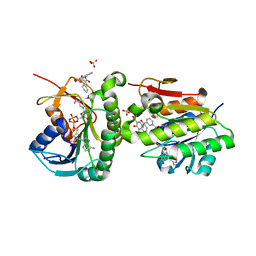 | | Crystal structure of human Biliverdin IX-beta reductase B with Pyrantel Pamoate (PPA) | | Descriptor: | 4-[(3-carboxy-2-oxidanyl-naphthalen-1-yl)methyl]-3-oxidanyl-naphthalene-2-carboxylic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7YYH
 
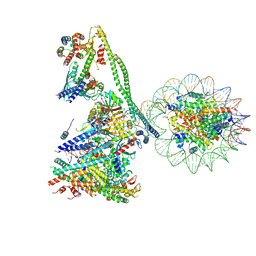 | | Structure of the human CCANdeltaT CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7ER6
 
 | | Crystal structure of human Biliverdin IX-beta reductase B | | Descriptor: | Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ERC
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Deferasirox (ICL) | | Descriptor: | 4-[3,5-bis(2-hydroxyphenyl)-1,2,4-triazol-1-yl]benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7ERA
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Olsalazine Sodium (OSS) | | Descriptor: | 5-[(E)-(3-carboxy-4-oxidanyl-phenyl)diazenyl]-2-oxidanyl-benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | Deposit date: | 2021-05-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
