8R2C
 
 | | Crystal structure of the Vint domain from Tetrahymena thermophila | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, von willebrand factor type A (VWA) domain was originally protein | | Authors: | Iwai, H, Beyer, H.M, Johannson, J.E, Li, M, Wlodawer, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of the Vint domain from Tetrahymena thermophila suggests a ligand-regulated cleavage mechanism by the HINT fold.
Febs Lett., 598, 2024
|
|
6ACJ
 
 | |
6R8T
 
 | | Crystal structure of aIF2gamma subunit I181T from archaeon Sulfolobus solfataricus complexed with GDPCP | | Descriptor: | FORMIC ACID, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Kravchenko, O, Arkhipova, V, Gabdulkhakov, A, Stolboushkina, E, Nikonov, O, Garber, M, Nikonov, S. | | Deposit date: | 2019-04-02 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of aIF2gamma subunit I181T from archaeon Sulfolobus solfataricus complexed with GDPCP
To Be Published
|
|
6RAA
 
 | | CLK1 Kinase domain with bound imidazopyridin inhibitor TP003 | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-methyl-1-(4-methylpiperazin-1-yl)-1-oxidanylidene-propan-2-yl]-~{N}-(6-pyridin-4-ylimidazo[1,2-a]pyridin-2-yl)benzamide, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Arrowsmith, C, Knapp, S, Bountra, C, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-05 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CLK1 Kinase domain with bound imidazopyridin inhibitor TP003
To Be Published
|
|
8QOA
 
 | | Structure of SecM-stalled Escherichia coli 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S16, ... | | Authors: | Gersteuer, F, Morici, M, Wilson, D.N. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | The SecM arrest peptide traps a pre-peptide bond formation state of the ribosome.
Nat Commun, 15, 2024
|
|
8R02
 
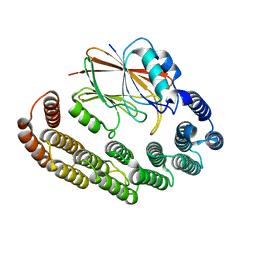 | | Crystal structure of the retromer complex VPS29/VPS35 with the ligand bis-1,3-phenyl guanylhydrazone, 2a | | Descriptor: | Bis-1,3-phenyl guanylhydrazon, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Milani, M, Fagnani, E. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stabilization of the retromer complex: Analysis of novel binding sites of bis-1,3-phenyl guanylhydrazone 2a to the VPS29/VPS35 interface.
Comput Struct Biotechnol J, 23, 2024
|
|
6RC4
 
 | |
6RCK
 
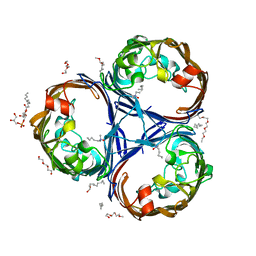 | | Crystal structure of the OmpK36 GD insertion chimera from Klebsiella pneumonia | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Beis, K, Romano, M, Kwong, J. | | Deposit date: | 2019-04-11 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | OmpK36-mediated Carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo.
Nat Commun, 10, 2019
|
|
8QZK
 
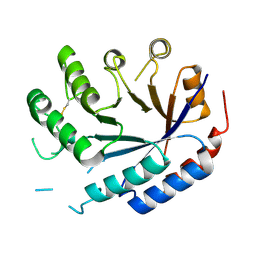 | | Catalytic core of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) concieved by deep network hallucination: dEngBF4 Hexagonal form | | Descriptor: | ENDO-ALPHA-N-ACETYLGALACTOSAMINIDASE | | Authors: | Aghajari, N, Hansen, A.L, Thiesen, F.F, Crehuet, R, Marcos, E, Willemoes, M. | | Deposit date: | 2023-10-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Carving out a Glycoside Hydrolase Active Site for Incorporation into a New Protein Scaffold Using Deep Network Hallucination.
Acs Synth Biol, 13, 2024
|
|
5ZWF
 
 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having a enone with a beta methyl group via conjugate addition reaction | | Descriptor: | (E,7R)-7-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-2-en-4-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
6A0K
 
 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, complex with panose | | Descriptor: | CALCIUM ION, Cyclic maltosyl-maltose hydrolase, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
8PPQ
 
 | | Tick-borne encephalitis virus Kuutsalo-14 prM3E3 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Protein prM | | Authors: | Anastasina, M, Domanska, A, Pulkkinen, L.I.A, Butcher, S.J. | | Deposit date: | 2023-07-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The structure of immature tick-borne encephalitis virus supports the collapse model of flavivirus maturation.
Sci Adv, 10, 2024
|
|
8POQ
 
 | |
6R7T
 
 | | Crystal Structure of human Melanoma-associated antigen B1 (MAGEB1) in complex with nanobody | | Descriptor: | Melanoma-associated antigen B1, anti MAGEB1 nanobody | | Authors: | Ye, M, Newman, J, Pike, A.C.W, Burgess-Brown, N, Cooper, C.D.O, Bountra, C, Arrowsmith, C, Edwards, A, Gileadi, O, von Delft, F. | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Crystal Structure of Melanoma-associated antigen B1 (MAGEB1) in complex with nanobody
To Be Published
|
|
6R83
 
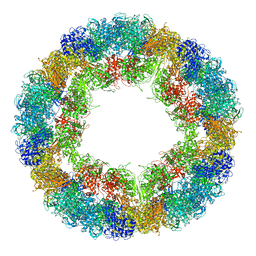 | | CryoEM structure and molecular model of squid hemocyanin (Todarodes pacificus , TpH) | | Descriptor: | Hemocyanin subunit 1 | | Authors: | Tanaka, Y, Kato, S, Stabrin, M, Raunser, S, Matsui, T, Gatsogiannis, C. | | Deposit date: | 2019-03-31 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryo-EM reveals the asymmetric assembly of squid hemocyanin.
Iucrj, 6, 2019
|
|
6A2J
 
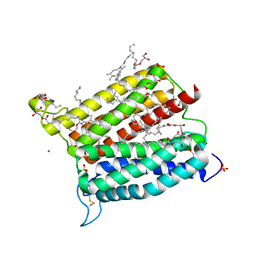 | | Crystal structure of heme A synthase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Heme A synthase, ... | | Authors: | Niwa, S, Takeda, K, Kosugi, M, Tsutsumi, E, Miki, K. | | Deposit date: | 2018-06-12 | | Release date: | 2018-11-21 | | Last modified: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of heme A synthase fromBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AAQ
 
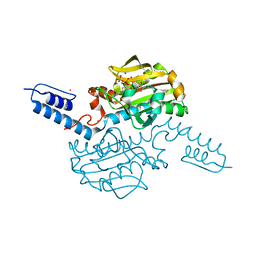 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with BCNLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N6-({[(1R,8S,9s)-bicyclo[6.1.0]non-4-yn-9-yl]methoxy}carbonyl)-L-lysine, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
8POR
 
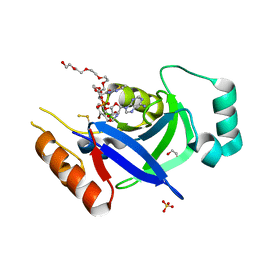 | |
6AB5
 
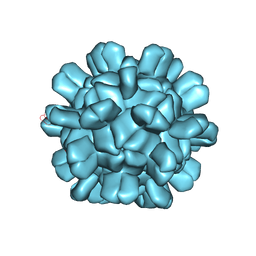 | | Cryo-EM structure of T=1 Penaeus vannamei nodavirus | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
6ABJ
 
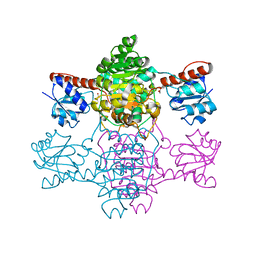 | | The apo-structure of D-lactate dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, D-lactate dehydrogenase (Fermentative) | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
8PFR
 
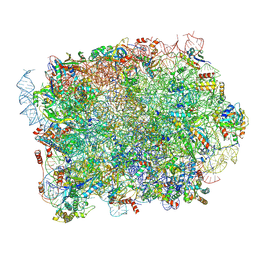 | | Mouse RPL39L integrated into the yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Mouse RPL39L integrated into the yeast 60S ribosomal subunit
To Be Published
|
|
6A5T
 
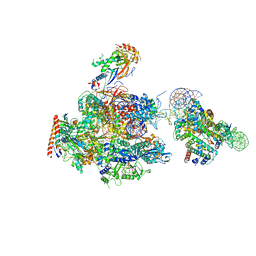 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6OLJ
 
 | |
8PIA
 
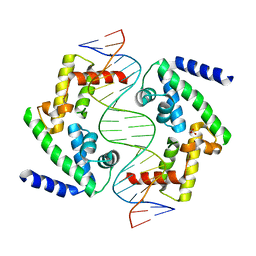 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA variant (P2 -181G>T) | | Descriptor: | Chains: E, Chains: F, GLYCEROL, ... | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
8PEM
 
 | |
