7OKX
 
 | | Structure of active transcription elongation complex Pol II-DSIF (SPT5-KOW5)-ELL2-EAF1 (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Vos, S.M, Dienemann, C, Ninov, M, Urlaub, H, Cramer, P. | | Deposit date: | 2021-05-18 | | Release date: | 2021-07-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric transcription stimulation by RNA polymerase II super elongation complex.
Mol.Cell, 81, 2021
|
|
1DGO
 
 | |
8UAZ
 
 | | Structure of Glutamyl-5'-O-adenosine phosphoramidate/RNase A | | Descriptor: | (2S)-2-{[(S)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]amino}pentanedioic acid (non-preferred name), DI(HYDROXYETHYL)ETHER, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2023-09-22 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Prolinyl Phosphoramidates of Nucleotides with Increased Reactivity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1DHG
 
 | | HG-SUBSTITUTED DESULFOREDOXIN | | Descriptor: | MERCURY (II) ION, PROTEIN (DESULFOREDOXIN) | | Authors: | Archer, M, Carvalho, A.L, Teixeira, S, Moura, I, Moura, J.J.G, Rusnak, F, Romao, M.J. | | Deposit date: | 1999-03-22 | | Release date: | 1999-07-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies by X-ray diffraction on metal substituted desulforedoxin, a rubredoxin-type protein.
Protein Sci., 8, 1999
|
|
8UIX
 
 | |
7OKY
 
 | | Structure of active transcription elongation complex Pol II-DSIF-ELL2-EAF1(composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Vos, S.M, Dienemann, C, Ninov, M, Urlaub, H, Cramer, P. | | Deposit date: | 2021-05-18 | | Release date: | 2021-07-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Allosteric transcription stimulation by RNA polymerase II super elongation complex.
Mol.Cell, 81, 2021
|
|
1DC4
 
 | | STRUCTURAL ANALYSIS OF GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE FROM ESCHERICHIA COLI: DIRECT EVIDENCE FOR SUBSTRATE BINDING AND COFACTOR-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Yun, M, Park, C.-G, Kim, J.-Y, Park, H.-W. | | Deposit date: | 1999-11-04 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli: direct evidence of substrate binding and cofactor-induced conformational changes.
Biochemistry, 39, 2000
|
|
7OL0
 
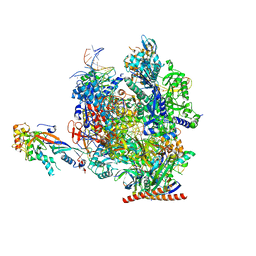 | | Structure of active transcription elongation complex Pol II-DSIF (SPT5-KOW5) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Vos, S.M, Dienemann, C, Ninov, M, Urlaub, H, Cramer, P. | | Deposit date: | 2021-05-18 | | Release date: | 2021-07-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Allosteric transcription stimulation by RNA polymerase II super elongation complex.
Mol.Cell, 81, 2021
|
|
8UB2
 
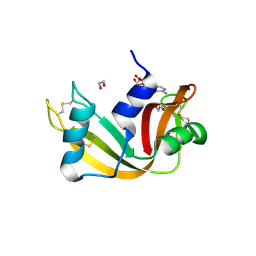 | | Structure of Adenosine monophosphate/RNase A | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2023-09-22 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Prolinyl Phosphoramidates of Nucleotides with Increased Reactivity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
3OP4
 
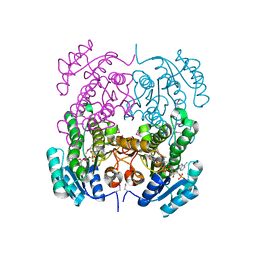 | | Crystal structure of putative 3-ketoacyl-(acyl-carrier-protein) reductase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with NADP+ | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hou, J, Chruszcz, M, Onopriyenko, O, Grimshaw, S, Porebski, P, Zheng, H, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
7SUU
 
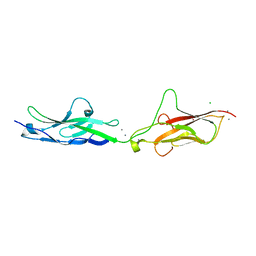 | |
1DG4
 
 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK IN THE APO FORM | | Descriptor: | DNAK | | Authors: | Pellecchia, M, Montgomery, D.L, Stevens, S.Y, Van der Kooi, C.W, Feng, H, Gierasch, L.M, Zuiderweg, E.R.P. | | Deposit date: | 1999-11-23 | | Release date: | 1999-12-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insights into substrate binding by the molecular chaperone DnaK.
Nat.Struct.Biol., 7, 2000
|
|
7SX6
 
 | |
8UGM
 
 | |
7SX7
 
 | |
3NO8
 
 | | Crystal structure of the PHR domain from human BTBD2 Protein | | Descriptor: | BTB/POZ domain-containing protein 2, GLYCEROL, SULFATE ION | | Authors: | Sampathkumar, P, Miller, S, Rutter, M, Bain, K, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the PHR domain from human BTBD2 Protein
To be Published
|
|
7ORE
 
 | | Crystal structure of JNK3 in complex with light-activated covalent inhibitor MR-II-249 with both non-covalent and covalent binding modes (compound 4) | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[(5Z)-9-[[4-[5-(4-fluorophenyl)-3-methyl-2-methylsulfanyl-imidazol-4-yl]pyridin-2-yl]amino]-11,12-dihydrobenzo[c][1,2]benzodiazocin-2-yl]butanamide, Mitogen-activated protein kinase 10 | | Authors: | Chaikuad, A, Reynders, M, Trauner, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Controlling the Covalent Reactivity of a Kinase Inhibitor with Light.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1DLB
 
 | | HELICAL INTERACTIONS IN THE HIV-1 GP41 CORE REVEALS STRUCTURAL BASIS FOR THE INHIBITORY ACTIVITY OF GP41 PEPTIDES | | Descriptor: | HIV-1 ENVELOPE GLYCOPROTEIN GP41 | | Authors: | Shu, W, Liu, J, Ji, H, Rading, L, Jiang, S, Lu, M. | | Deposit date: | 1999-12-09 | | Release date: | 1999-12-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Helical interactions in the HIV-1 gp41 core reveal structural basis for the inhibitory activity of gp41 peptides.
Biochemistry, 39, 2000
|
|
7OMA
 
 | | Thosea asigna virus RdRP domain elongation complex | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, RNA (5'-R(P*AP*AP*AP*UP*UP*UP*U)-3'), ... | | Authors: | Ferrero, D.S, Falqui, M, Verdaguer, N. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of a Non-Canonical RdRP in Action.
Viruses, 13, 2021
|
|
7OM9
 
 | |
6I9Y
 
 | | The 2.14 A X-ray crystal structure of Sporosarcina pasteurii urease in complex with Au(I) ions | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, HYDROXIDE ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-11-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Inhibition Mechanism of Urease by Au(III) Compounds Unveiled by X-ray Diffraction Analysis.
Acs Med.Chem.Lett., 10, 2019
|
|
2OV2
 
 | | The crystal structure of the human RAC3 in complex with the CRIB domain of human p21-activated kinase 4 (PAK4) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ugochukwu, E, Yang, X, Elkins, J.M, Burgess-Brown, N, Bunkoczi, G, Debreczeni, J.E.D, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the human RAC3 in complex with the CRIB domain of human p21-activated kinase 4 (PAK4)
To be Published
|
|
7OM6
 
 | |
1DDK
 
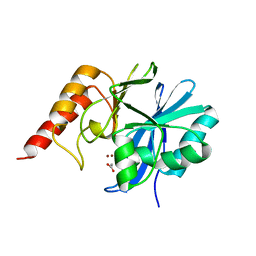 | | CRYSTAL STRUCTURE OF IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, ZINC ION | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
8VWL
 
 | |
