7OLX
 
 | | MerTK kinase domain with type 1.5 inhibitor containing a tri-methyl pyrazole group | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer, ~{N}-[[3-[4-[(dimethylamino)methyl]phenyl]imidazo[1,2-a]pyridin-6-yl]methyl]-~{N}-methyl-5-[3-methyl-5-(1,3,5-trimethylpyrazol-4-yl)pyridin-2-yl]-1,3,4-oxadiazol-2-amine | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
7OZM
 
 | | Crystal Structure of mtbMGL K74A (Closed Cap Conformation) | | Descriptor: | ISOPROPYL ALCOHOL, Monoacylglycerol lipase | | Authors: | Grininger, C, Aschauer, P, Pavkov-Keller, T, Oberer, M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Changes in the Cap of Rv0183/mtbMGL Modulate the Shape of the Binding Pocket.
Biomolecules, 11, 2021
|
|
7P0Y
 
 | | Crystal Structure of mtbMGL K74A (Substrate Analog Complex) | | Descriptor: | 1-[butyl(fluoranyl)phosphoryl]oxyhexadecane, Monoacylglycerol lipase | | Authors: | Grininger, C, Aschauer, P, Pavkov-Keller, T, Oberer, M. | | Deposit date: | 2021-06-30 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Changes in the Cap of Rv0183/mtbMGL Modulate the Shape of the Binding Pocket.
Biomolecules, 11, 2021
|
|
4RBY
 
 | |
4RC0
 
 | |
7OLS
 
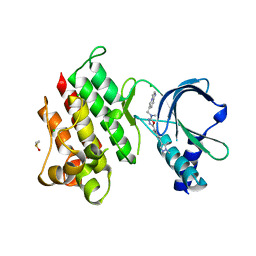 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl pyrazole group | | Descriptor: | 5-[4-(1,5-dimethylpyrazol-4-yl)-2-methyl-phenyl]-~{N}-(imidazo[1,2-a]pyridin-6-ylmethyl)-~{N}-methyl-1,3,4-oxadiazol-2-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
7OLV
 
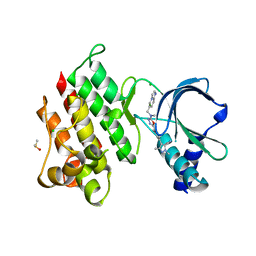 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl, cyano pyrazole group | | Descriptor: | 4-[4-[5-[imidazo[1,2-a]pyridin-6-ylmethyl(methyl)amino]-1,3,4-oxadiazol-2-yl]-3-methyl-phenyl]-2,5-dimethyl-pyrazole-3-carbonitrile, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
4QQK
 
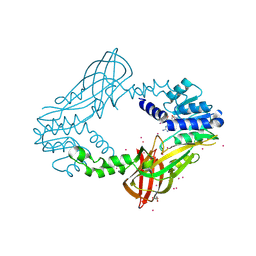 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) with GMS | | Descriptor: | (5S)-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}-N~6~-carbamimidoyl-L-lysine, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
6QSC
 
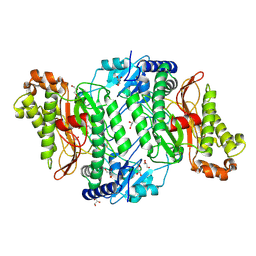 | | Crystal Structure of Arg470His mutant of Human Prolidase with Mn ions and GlyPro ligand | | Descriptor: | GLYCEROL, GLYCINE, MANGANESE (II) ION, ... | | Authors: | Wilk, P, Wator, E, Weiss, M.S. | | Deposit date: | 2019-02-20 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.569 Å) | | Cite: | Structural analysis of new compound heterozygous variants in PEPD gene identified in a patient with Prolidase Deficiency diagnosed by exome sequencing.
Genet Mol Biol, 44, 2021
|
|
6R02
 
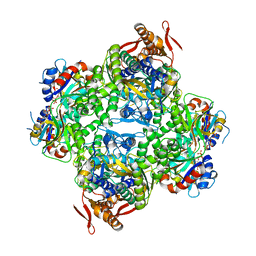 | | Psychrobacter arcticus ATP phosphoribosyltransferase bound to histidine and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, ... | | Authors: | Alphey, M.S, da Silva, R.G, Thomson, C.M. | | Deposit date: | 2019-03-12 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mapping the Structural Path for Allosteric Inhibition of a Short-Form ATP Phosphoribosyltransferase by Histidine.
Biochemistry, 58, 2019
|
|
1DKH
 
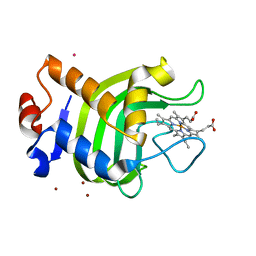 | | CRYSTAL STRUCTURE OF THE HEMOPHORE HASA, PH 6.5 | | Descriptor: | HEME-BINDING PROTEIN A, PROTOPORPHYRIN IX CONTAINING FE, SAMARIUM (III) ION, ... | | Authors: | Arnoux, P, Haser, R, Izadi-Pruneyre, N, Lecroisey, A, Czjzek, M. | | Deposit date: | 1999-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Functional aspects of the heme bound hemophore HasA by structural analysis of various crystal forms.
Proteins, 41, 2000
|
|
7OTD
 
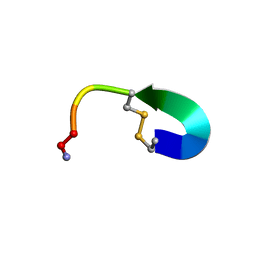 | | Oxytocin NMR solution structure | | Descriptor: | AMINO GROUP, COPPER (II) ION, UNK-TYR-ILE-GLN-ASN-CYS-PRO-LEU-GLY | | Authors: | Shalev, D.E, Alshanski, I, Yitzchaik, S, Hurevich, M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-13 | | Method: | SOLUTION NMR | | Cite: | Determining the structure and binding mechanism of oxytocin-Cu 2+ complex using paramagnetic relaxation enhancement NMR analysis.
J.Biol.Inorg.Chem., 26, 2021
|
|
5Y0D
 
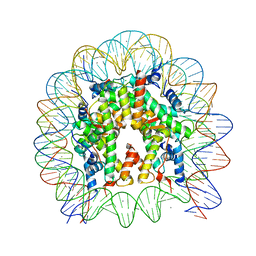 | | Crystal Structure of the human nucleosome containing the H2B E76K mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | Deposit date: | 2017-07-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
6R0H
 
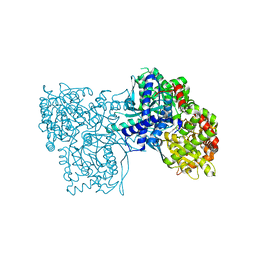 | | Glycogen Phosphorylase b in complex with 3 | | Descriptor: | 3-(4-fluorophenyl)-~{N}-[(2~{R},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]benzamide, Glycogen phosphorylase, muscle form, ... | | Authors: | Tsagkarakou, S.A, Koulas, M.S, Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-10 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High Consistency of Structure-Based Design and X-Ray Crystallography: Design, Synthesis, Kinetic Evaluation and Crystallographic Binding Mode Determination of Biphenyl-N-acyl-beta-d-Glucopyranosylamines as Glycogen Phosphorylase Inhibitors.
Molecules, 24, 2019
|
|
6R0I
 
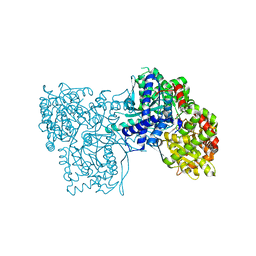 | | Glycogen Phosphorylase b in complex with 4 | | Descriptor: | Glycogen phosphorylase, muscle form, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Koulas, M.S, Tsagkarakou, S.A, Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-17 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High Consistency of Structure-Based Design and X-Ray Crystallography: Design, Synthesis, Kinetic Evaluation and Crystallographic Binding Mode Determination of Biphenyl-N-acyl-beta-d-Glucopyranosylamines as Glycogen Phosphorylase Inhibitors.
Molecules, 24, 2019
|
|
7OVH
 
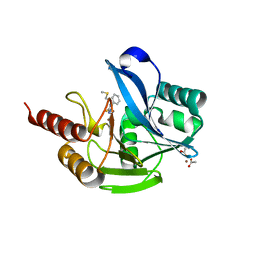 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in Complex with compound 14 (JMV-6931) | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, UNKNOWN LIGAND, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Sannio, F, Marcoccia, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,2,4-Triazole-3-thione compounds with a 4-ethyl alkyl/aryl sulfide substituent are broad-spectrum metallo-beta-lactamase inhibitors with re-sensitization activity.
Eur.J.Med.Chem., 226, 2021
|
|
7OVE
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in Complex with compound 10 (JMV-7210) | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, UNKNOWN LIGAND, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Sannio, F, Mangani, S, Docquier, J.D, Pozzi, C, Marcoccia, F. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | 1,2,4-Triazole-3-thione compounds with a 4-ethyl alkyl/aryl sulfide substituent are broad-spectrum metallo-beta-lactamase inhibitors with re-sensitization activity.
Eur.J.Med.Chem., 226, 2021
|
|
7OVF
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in Complex with compound 8 (JMV-7207) | | Descriptor: | 4-[2-[(4-fluorophenyl)methylsulfanyl]ethyl]-3-phenyl-1H-1,2,4-triazole-5-thione, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Sannio, F, Marcoccia, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1,2,4-Triazole-3-thione compounds with a 4-ethyl alkyl/aryl sulfide substituent are broad-spectrum metallo-beta-lactamase inhibitors with re-sensitization activity.
Eur.J.Med.Chem., 226, 2021
|
|
7P9C
 
 | | Escherichia coli type II L-asparaginase | | Descriptor: | L-asparaginase 2 | | Authors: | Maggi, M, Scotti, C. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Revealing Escherichia coli type II L-asparaginase active site flexible loop in its open, ligand-free conformation.
Sci Rep, 11, 2021
|
|
7CFO
 
 | | Crystal structure of human RXRalpha ligand binding domain complexed with CBTF-EE. | | Descriptor: | 1-[3-(2-ethoxyethoxy)-5,5,8,8-tetramethyl-6,7-dihydronaphthalen-2-yl]-2-(trifluoromethyl)benzimidazole-5-carboxylic acid, GLYCEROL, Retinoic acid receptor RXR-alpha | | Authors: | Watanabe, M, Fujihara, M, Motoyama, T, Kawasaki, M, Yamada, S, Takamura, Y, Ito, S, Makishima, M, Nakano, S, Kakuta, H. | | Deposit date: | 2020-06-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a "Gatekeeper" Antagonist that Blocks Entry Pathway to Retinoid X Receptors (RXRs) without Allosteric Ligand Inhibition in Permissive RXR Heterodimers.
J.Med.Chem., 64, 2021
|
|
7OR2
 
 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 4) | | Descriptor: | 5-methyl-1-phenyl-pyrazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-04 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OSQ
 
 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 5-methyl-1-phenyl-1,2,3-triazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Mayol-Llinas, J, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
3D8B
 
 | | Crystal structure of human fidgetin-like protein 1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fidgetin-like protein 1 | | Authors: | Karlberg, T, Wisniewska, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Welin, M, Wikstrom, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human fidgetin-like protein 1.
To be Published
|
|
7OXD
 
 | |
8GW4
 
 | | SARS-CoV-2 Mpro 1-302/C145A in complex with peptide 8-1 | | Descriptor: | Replicase polyprotein 1ab, peptide 8-1 | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
